Back to Journals » OncoTargets and Therapy » Volume 11
MicroRNA-384 inhibits proliferation migration and invasion of glioma by targeting at CDC42
Authors Gu G, Wang L, Zhang J, Wang H, Tan T, Zhang G
Received 27 February 2018
Accepted for publication 18 April 2018
Published 16 July 2018 Volume 2018:11 Pages 4075—4085
DOI https://doi.org/10.2147/OTT.S166747
Checked for plagiarism Yes
Review by Single anonymous peer review
Peer reviewer comments 2
Editor who approved publication: Dr XuYu Yang
Gengshi Gu,1,* Li Wang,1,* Junchen Zhang,1 Hao Wang,2 Tan Tan,1 Guangning Zhang1
1Department of Neurosurgery, Affiliated Hospital of Jining Medical University, Jining, Shandong 272000, People’s Republic of China; 2Department of Clinical Laboratory, Chinese People’s Liberation Army General Hospital, Beijing, 100700, People’s Republic of China
*These authors contributed equally to this work
Background: Accumulative evidence indicated that microRNAs (miRNAs) play a critical role in carcinogenesis and biological behaviors of glioma. Further bio-molecular mechanisms of miRNAs in glioma cells remain largely unknown, which can contribute to novel therapeutic strategy.
Methods: In the present study, we detected the expression level of miR-384 by RT-PCR and Western blot. Meanwhile, Gain and loss function assay of miR-384 by transfection of miR-384 mimics and inhibitor. Moreover, wild and mutant psiCHECK-2-CDC42-3’-UTR luciferase reporter vectors were constructed and transfected into glioma cells with miR-384 mimics or miR-NC.
Results: miR-384 was dramatically down-regulated in human glioma tissues. It was also demonstrated that miR-384 significantly inhibited proliferation, migration and invasion of glioma cells. Cell division cycle 42 (Cdc42) was a direct target of miR-384 according to results of RT-PCR and Western blotting.
Conclusion: Our research demonstrated that miR-384 exerted an inhibitory effect on proliferation, migration and invasion of glioma via suppressing the expression of CDC42, meaning that miR-384 may be regarded as a potential target in the treatment of glioma.
Keywords: miR-384, CDC42, glioma, proliferation, invasion
Introduction
Glioma is the most common primary malignant tumor of the central nervous system in adults with high morbidity and mortality, which causes serious harm to human health.1 Despite a significant therapeutic improvement over the past few decades, the prognosis and survival of patients with gliomas are still unfavorable, and the high aggressiveness of glioma dramatically limits its overall therapeutic efficacy. Specifically, glioblastoma multiforme is the most malignant glioma.2 Therefore, investigations on a further molecular mechanism of glioma invasion and more appropriate therapeutic target are of great significance.
MicroRNAs (miRNAs) are a group of cytoplasmic non-coding RNAs that contain between 19 and 24 nucleotides in length.3 It was found that miRNAs play an important role in many biological behaviors such as cell proliferation, differentiation, apoptosis, tumorigenesis, migration, invasion and neuronal development. They function as negative regulators of gene expression by binding to the 3′-untranslated region (3′-UTR) of miRNAs of the target gene, resulting in miRNA degradation and inhibition of protein translation.4 Depending on the target gene, miRNAs can function as a tumor suppressor or oncogenes, inhibiting translation of proteins primarily by complementarily combining with miRNAs, thereby exerting their regulatory role at the post-transcriptional level. In recent years, miRNAs have become important regulators of human cancer. The expression of miRNA has been used as an indicator of diagnosis, progression, prognosis and therapeutic efficacy of cancer.5 More and more studies have shown that miRNAs play an important role in the development of neurological tumors.6–8 MiR-384 is abnormally expressed in many malignant tumors such as thyroid cancer, pancreatic cancer, hepatocellular carcinoma (HCC) and colorectal cancer.9–12
Cell division cycle 42 (CDC42), as a member of the Rho protein family, is a guanosine triphosphatase and responds to extracellular signals.13 CDC42 plays a switch function to activate or inactive downstream molecules by interacting with GTP or GDP, respectively.14 However, abnormal activation or mutants of CDC42, such as the constitutively active mutants CDC42Q61L and CDC42G12V, exist in many cancers. CDC42F28L, a spontaneous accelerated circulating mutant of GDP-GTP binding state, was capable of inducing colony formation or non-anchoring dependent growth of immortalized fibroblasts.15–17 Among other Rho proteins, CDC42 promotes malignant transformation by altering cellular metabolism.18
Previous studies19–21 have shown that CDC42 also plays a critical role in the development of glioma. Tang et al22 revealed that LRRC4 in glioma cell lines positively regulates the expression of miR-185, while miR-185 participates in tumor growth and metastasis by negatively regulating the expression of CDC42. This study will explore the role of miR-384 in proliferation, migration and invasion of glioma by regulating the downstream gene CDC42.
Materials and methods
Clinical samples and cell culture
This study was carried out in accordance with the recommendations of institutional review board of Affiliated Hospital of Jining Medical University with written informed consent from all subjects. All donors signed the informed consent in accordance with the Declaration of Helsinki. The protocol was approved by the institutional review board of Affiliated Hospital of Jining Medical University.
A total of 48 glioma samples were obtained from patients undergoing surgical resection for diffuse gliomas at Department of Neurosurgery of Affiliated Hospital of Jining Medical University from December 2014 to September 2015. Clinical and pathological materials were collected and statistically analyzed (Table 1). In total, 24 non-neoplastic brain samples were obtained from patients with traumatic brain injury. These samples were immediately kept in liquid nitrogen right after resected for subsequent investigations, none of which had been dealt with pre-operational radiotherapy or chemotherapy. Diagnoses were made by an experienced neuropathologist according to 2016 World Health Organization (WHO) classification of central nervous system (CNS) tumor.23 Grades I and II were defined as low grade, and grades III and IV were defined as high grade.
  | Table 1 Association between miR-384 expression and clinical and pathological parameters in 48 patients with glioma |
The glioma cell lines including U251, U87, T98G, SNB19, U373, A172 and normal human astrocytes (NHAs) were obtained from the Cell Bank of Shanghai Life Academy of Science. These cells were cultured with DMEM containing 100 U/mL penicillin, 100 μg/mL streptomycin and 10% fetal bovine serum (FBS) in a 5% CO2 atmosphere at 37°C.
Real-time quantitative polymerase chain reaction (RT-qPCR)
According to manufacturer’s instructions, total RNA was extracted using Trizol reagent (Invitrogen, Thermo Fisher Scientific, Waltham, MA, USA) for both cell lines and patients’ samples. The expression of miR-384 and CDC42 was detected by SYBR Premix Ex Taq system (Takara, Madison, WI, USA) according to the manufacturer’s instructions. RT-PCR reactions were performed in triplicates using the following cycling parameters: initial denaturation at 95°C for 30 s, 40 cycles of denaturation at 95°C for 5 s and annealing at 60°C for 34 s. We assessed the RNA expression according to relative quantification using the 2−ΔΔCt method described previously to determine the fold change in the expression.24 The U6 small nuclear RNA and GAPDH mRNA were used as internal controls for miR-384 and CDC42 mRNA, respectively. The sequences of primers for RT-PCR were as follows: CDC42 forward primer: 5′-GAAGGCTGTCAAGTATGTGG-3′, reverse primer: 5′-CTCTTCTTCGGTTCTGGAGG-3′, GAPDH forward primer: 5′-ACCCAGAAGACTGTGGATGG-3′, reverse primer: 5′-CACATTGGGGGTAGGAACAC-3′, U6 small nuclear RNA forward primer: 5′-GTGCTCGCTTCGGCAGCACATATAC-3′, reverse primer: 5′-AAAAATATGGAACGCTCACGAATTTG-3′. All data were expressed as mean ± SD and as representative of an average of three measurements.
miRNA transfection, CDC42 silencing and overexpression
The miR-384 mimics, miR-384 inhibitor, miR negative control (miR-NC) and inhibitor-NC, as the relative controls, were designed and provided by GenePharma (Shanghai, People’s Republic of China). The following sequences were used: miR-384: 5′-AUUCCUAGAAAUUGUUCAUA-3′, miR-NC: 5′-UUCUCCGAACGUGUCACGU-3′, miR-384 inhibitor: 5′-UAUGAACAAUUUCUAGGAAU-3′ and inhibitor-NC: 5′-CAGUACUUUUGUGUAGUACAA-3′. The miR-384 inhibitor, a single-stranded RNA nucleotide chain, can specifically target at miR-384 to degrade it. Lipofectamine 2000 (Invitrogen, Thermo Fisher Scientific) was used to transfect cells with miR-384 mimics and inhibitor according to the manufacturer’s protocols. Transfection efficiency was detected by RT-qPCR and Western blot analysis. siRNA (5′-GACTACGACCGCTAAGTTA-3′) targeting at CDC42 was designed and purchased from Shanghai Sangon Co., Ltd. (Shanghai, People’s Republic of China) and transfection were performed using Lipofectamine 2000 (Invitrogen, Thermo Fisher Scientific) according to manufacturer’s instructions to transfect U251 cells with siRNA. Additionally, we transfected U251 cells with pcDNA3.1-Cdc42 provided by Youbo Biological Technology Co., Ltd. (Beijing, People’s Republic of China) to overexpress CDC42. Afterward, proliferation and invasion of both wild-type (wt) and CDC42 overexpressed glioma cells transfected with miR-NC or miR-384 were investigated by cell counting kit-8 (CCK-8; Dojindo, Kumamoto, Japan) assay and transwell assay, respectively.
Cell proliferation assay by CCK-8
Cell proliferation assay was performed using CCK-8 (Dojindo) according to manufacturer’s instruction. Briefly, 1×104 cells/well of U251, U87, T98G and A172 were seeded into 96-well plates. Overnight culture after cell adhesion, miR-384 mimics or miR-NC was added into culture media to transfect cells at a final concentration of 50 nM/uL. Then 0, 24, 48, 72 and 96 h after transfection, CCK-8 was added into culture media at a final concentration of 10 uL/100 uL followed by incubation at 37°C for 2 h. The absorbance was immediately detected at a wavelength of 450 nm (630 nm as reference) on a micro-plate analyzer (Bio-Rad, Hercules, CA, USA).
Colony formation assay
The colony formation assay was conducted as previously described.25 Briefly, 6-well plate was covered with 2 mL 1:1 mixed 1.2% agar and 2× dulbecco’s modified eagle medium (DEME) at room temperature for solidification. Then, 2 mL mixture of 0.7% agar and 2× DEME (1:1 mixed) containing 100 uL 5×104/mL U251 cells that were transfected with miR-384 mimics, miR-NC, miR-384 inhibitor or inhibitor-NC for 48 h was added into wells. After incubation in 5% CO2 and 37°C for 14 days, they were fixed with 10% formaldehyde and stained with 0.1% crystal violet (Sigma-Aldrich Co.). Colony number was counted under a microscope and presented as a histogram. The average number of colonies was calculated at 5 randomly views in three replicative wells. The experiment was repeated for three independent times.
Wound-healing assay
In total, 8×104 U251 cells were seeded onto 6-well plates and incubated for 24 h until adhesion, and then cells were transfected with miR-384 or miR-NC using Lipofectamine 2000 (Invitrogen, Thermo Fisher Scientific). When cells reached 90% confluence after transfection for about 48 h, wounds were scratched in the center of confluent cells with a pipette tip. Then, the healing was observed and photographed under microscope, and cell migration distances were also calculated at 12 and 24 h after scratching. Each experiment was repeated for at least three times.
Transwell invasion assay
For invasion assay of glioma cells, 24-well transwell inserts (Corning Costar, Cambridge, MA, USA) were coated on their bottom by Matrigel Basement Membrane Matrix (BD Biosciences, San Jose, CA, USA). After transfecting with miR-384, miR-NC, miR-384 inhibitor or inhibitor-NC for 48 h, 5×104 U251 cells were seeded onto upper chamber within 200 uL serum-free DEME, while 500 uL DEME with 10% FBS as chemoattractants were added into lower chambers. After incubation for 48 h, the non-invasive cells on upper side were washed away with PBS for three times. The membranes were fixed with 4% paraformaldehyde and then stained with 0.1% crystal violet (Sigma-Aldrich Co.). The average number of invasive cells were observed and counted under a microscope from 5 random views of each three replicate wells.
Western blot analysis
For extraction of total proteins, 1×106 U251 and U87 cells were seeded onto 6-well plates; transfected with miR-384, miR-NC, miR-384 inhibitor or inhibitor-NC; and then incubated for 48 h until about 90% confluence. Then, cells were lysed by radioimmunoprecipitation assay (RIPA) buffer with phenylmethylsulfonyl fluoride (PMSF), a protease inhibitor, and supernatants were extracted after centrifugation. Concentrations of protein were detected using a bicinchoninic acid (BCA) kit (Thermo Fisher Scientific). Protein extracts were separated on sodium dodecyl sulfate-polyacrylamide gels and transferred onto polyvinylidene difluoride membranes (Millipore, Bedford, MA, USA). After blocking with 5% non-fat milk, the membrane was incubated with anti-CDC42 and anti-GAPDH antibodies (Cell Signaling Technology, Danvers, MA, USA) at 4°C overnight. The membrane was then blotted with secondary antibodies for 1 h at room temperature and visualized by a chemiluminescence system (Millipore).
Luciferase reporter assay
The miR-384 binding site located at 1329–1334 bp in CDC42 3′-UTR. The cDNA of wide-type or mutant CDC42 was synthesized and integrated into a psiCHECK-2 dual-luciferase vector to form the psiCHECK-2-CDC42-3′-UTR-wild type (CDC42-wt) or psiCHECK-2-CDC42-3′-UTR-mutant (CDC42-mut) reporter vector. For luciferase reporter assay, 1×105 U251, U87 and T98G cells/well were seeded onto 24-well plates and incubated for 24 h. The luciferase reporter gene plasmids, including wide-type or mutant psiCHECK-2-CDC42-3′-UTR, were transfected into cells using Lipofectamine 2000 (Invitrogen, Thermo Fisher Scientific) according to the manufacturer’s instruction. Specifically, 800 ng psiCHECK-2-CDC42-3′-UTR vectors were co-transfected with 50 pmol miR-384 mimics, miR-384 inhibitors, inhibitor-NC or miR-NC respectively. After 24 h of incubation following transfection, the luciferase activity was measured using a dual luciferase reporter assay kit (Promega, Madison, WI, USA) according to the manufacturer’s protocol. All experiments were conducted for at least three times.
Statistical analysis
Data in the present study were shown as mean ± SD from three independent replicative assays and analyzed using SPSS 18.0 software (SPSS Inc., Chicago, IL, USA) and GraphPad Prism 6 (GraphPad Software Inc., San Diego, CA, USA). A value of p<0.05 means statistically significant.
Results
MiR-384 was downregulated in glioma cell lines and human glioma tissues
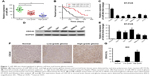
The role of miR-384 in tumorigenesis of glioma was investigated in both normal and glioma cell lines and tissues by quantitative RT-PCR. Results showed that the expression level of miR-384 was significantly downregulated in glioma cell lines, including U251, U87, T98G, SNB19, U373 and A172, in comparison to that of NHAs (p<0.05, Figure 1C). Meanwhile, the expression of miR-384 was remarkably decreased in human glioma tissue compared with that in normal human brain tissue in a grade-dependent manner, which demonstrated that high-grade glioma tissue, classified according to 2016 WHO classification of CNS tumor, manifested much lower level of miR-384 than low-grade glioma tissue (p<0.05, Figure 1A). Then, we reviewed and analyzed clinical materials, and investigated the correlation between miR-384 level in glioma tissue of patients and overall survival using the Kaplan-Meier survival curve. Patients were separated into the low miR-384 group and the high miR-384 group referring to median level in all patients. The results revealed that patients with the high miR-384 level obviously had a significant good prognosis than the low miR-384 group (p<0.01, Figure 1B). We detected the expression of CDC42 in several glioma cell lines as well as NHAs by RT-PCR and Western blot analysis. It turns out that mRNA and protein of CDC42 were significantly higher in glioma cells than in the control group (Figure 1D and E). In addition, immunohistochemistry assay was performed to investigate the expression of CDC42 in glioma tissue and normal brain tissue. In line with the results of cell lines, normal brain tissues exhibited remarkably lower expression level of CDC42 compared with glioma tissues (Figure 1F and G).
Inhibiting proliferation of glioma cell lines by miR-384
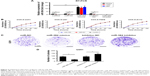
The expressions of miR-384 in NHAs, U251 and U87 transfected with miR-NC, miR-384 mimics, inhibitor-NC or miR-384 inhibitor were detected by RT-PCR. As a result, the expression level of miR-384 significantly increased in all miR-384 mimics-transfected cells (Figure 2A). Further investigations about the role of miR-384 in the proliferation of glioma cells were carried out. MiR-384 mimics and inhibitor and their controls were transfected into glioma cells for upregulating and downregulating its expression. First, the results of CCK-8 assay indicated that miR-384 remarkably inhibited the proliferation of U251, U87, T98G and A172 (Figure 2B). Additionally, colony formation assay of U251 was performed. Expression of miR-384 was increased and decreased by transfecting with miR-384 mimics and inhibitor. The results demonstrated that the number of formed colonies of miR-384 mimics group was significantly less than that of the miR-NC group. Correspondingly, miR-384 inhibitor not only degraded miR-384 in U251 cells but also greatly promoted formation of colonies manifested by crystal violet stain in comparison to that of inhibitor-NC group (p<0.05, Figure 2C and D).
Inhibiting migration and invasion of glioma cells by miR-384
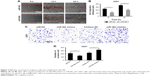
After transfection of miR-384 mimics for 48 h, migration of U251 cells was investigated by wound-scratch assay, in which the widths of wound between cells were observed and recorded over time. According to the results, it was observed that miR-384 significantly inhibited migration of U251 cells at 24 and 48 h compared with the miR-NC group (p<0.05, Figure 3A and B). Moreover, invasive capacities of glioma cells transfected with miR-384 mimics or miR-384 inhibitor were studied by transwell assay in which membranes were stained with crystal violet. According to the results, it was suggested that U251 cells transfected with miR-384 mimics exhibited a remarkable reduction in the number of positively stained cells that traversed the membrane in comparison to those with miR-NC (p<0.05). Furthermore, the number of positive stain U251 cells that were transfected with miR-384 inhibitor manifested significant increase compared with the control group (p<0.05, Figure 3C and D).
CDC42 is a potential target of miR-384 during its inhibition on glioma cells
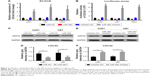
According to previous researches, CDC42, as a member of the Rho protein family, was a target gene of miR-384 in other carcinomas and of many other miRNAs as well.12,26–29 Therefore, we assumed that CDC42 might be the potential target of miR-384 during its inhibitory effects on proliferation, migration and invasion of glioma cells. mRNA of CDC42 in U251, U87 and T98G cells transfected with miR-384 mimics were significantly downregulated compared with transfecting miR-NC groups, while the suppression of CDC42 mRNA by miR-384 was abolished in cells transfected with miR-384 inhibitor (p<0.05, Figure 4A).
Luciferase reporter assay was used to further detect the influence of miR-384 on the expression of CDC42. As a result, in line with the consequence of RT-PCR, upregulation of miR-384 significantly suppressed the activity of reporter gene, psiCHECK-2-CDC42-3′-UTR, which was dramatically rescued by transfection of miR-384 inhibitor (p<0.05, Figure 4B).
Furthermore, proteins of CDC42 in U251 and U87 were detected by Western blotting. After transfection of miR-384 mimics and miR-NC for 48 h, total proteins were extracted and expressions of CDC42 were detected. The results showed that expressions of CDC42 were downregulated in miR-384 mimics-transfected group compared with those of miR-NC group (p<0.05, Figure 4C and D). On the contrary, CDC42 was upregulated in the miR-384 inhibitor-transfected group (p<0.05, Figure 4E and F). In order to compare the suppressive efficacy of miR-384 on CDC42, CDC42 knockout glioma cells were obtained by transfecting siRNA. Total proteins were extracted from wt and CDC42-/- and then expressions of CDC42 were measured by Western blot analysis. As a result, CDC42 was successfully silenced in siRNA-transfected group, while CDC42 was not completely suppressed by miR-384 in comparison to the silenced group (Figure 5A).
To further testify miR-384 targeting at CDC42-3′-UTR, CDC42-wt and CDC42-mut reporter vectors were constructed and used in luciferase reporter assay (Figure 5B). It turned out that, as expectedly, the relative luciferase activity of CDC42-wt with transfection of miR-384 group was greatly reduced in comparison to that of CDC42-wt with transfection of miR-NC (p<0.05, Figure 5C). However, in CDC42-mut cells transfected with miR-384, the luciferase activity was not suppressed by overexpression of miR-384 (p>0.05, Figure 5B). Afterward, proliferation and invasion of both wild type and CDC42 overexpressed glioma cells transfected with miR-NC or miR-384 were investigated by CCK-8 assay and transwell assay, respectively. It turns out that overexpression of CDC42 significantly rescued inhibitory effect of miR-384 on proliferation and invasion of glioma cells (Figure 5D and E).
Discussion
As the most common intracranial malignant tumor, glioma, with an incidence of 5/100,000, shows invasive growth. There might be tumor cells growing in normal brain tissues within 2 cm around the tumor. As malignant glioma has a very high invasive capacity, so even after resecting the whole tumor in an operation, its recurrence still cannot be avoided in the edge of the incision.30 Moreover, chemotherapy and radiotherapy not only failed to specifically kill glioma cells, leading to a poor therapeutic effect, but also may produce central nervous system toxicity and serious side effects. So the prognosis and the median survival are still far from expectation even with combined treatment. Given the abovementioned situation has not been fundamentally improved in nearly 30 years of clinical practice, gliomas are still the refractory disease in the field of neurosurgery.31 Therefore, to clarify its pathogenesis, the search for a new and effective therapeutic target remains critical.
In recent years, miRNAs have been the focus of research in life science. More and more evidence shows that miRNAs, as one way of epigenetic regulation of genes, play an important role in the development, progression and prognosis of human tumors, which have provided a new breakthrough in the genetic diagnosis and treatment, especially in the invasion and metastasis of tumors. As for the role of miRNAs in glioma, it has also been reported that multiple miRNAs, such as miR-106b-5p, miR-195 and miR-204, can promote glioma tumorigenesis by targeting multiple tumor suppressor genes.
In the present study, we detected the expression level of miR-384 in human glioma tissues and common glioma cell lines compared with those in normal human brain tissues and NHAs, respectively. As a result, miR-384 was dramatically reduced in glioma cells compared with normal ones, which indicated that miR-384 plays an an anti-oncogenic role. In addition, overall survivals of patients with low or high miR-384 level were followed up and recorded, and subsequently analyzed to investigate the relationship between overall survival of patients with glioma and expression level of miR-384. According to the results, it was observed that patients with high miR-384 showed a significantly longer survival than those with low miR-384.
Therefore, the effect of miR-384 on the proliferation of glioma cells was detected by CCK-8 at different time points, in which cell viabilities of multiple glioma cell lines including U251, U87, T98G and A172 were dramatically suppressed by transfection of miR-384 mimics in comparison to control groups. Correspondingly, Bai et al reported that downregulation of miR-384 expression was a common event in HCC, especially hepatitis B virus (HBV)-related HCC, which promotes proliferation, metastasis and lipogenesis.11 Likewise, effects of miR-384 mimics and miR-384 inhibitor on migration and invasion of glioma cells were investigated by wound-scratch assay and transwell assay, respectively; as a result, it was found that overexpression of miR-384 obviously inhibited migration and invasion of glioma cells.
However, miRNA usually works via targeting at downstream genes to regulate them. Therefore, according to previous researches, we know that CDC42 was a target of miR-384 in colorectal cancer and many other miRNAs.12,26–29 So, it is reasonable to assume that miR-384 inhibits proliferation, migration and invasion of glioma cells via targeting at CDC42. Then the results of RT-PCR and Western blot analysis showed that CDC42 was downregulated in glioma cells transfected with miR-384 mimics and this inhibition was rescued in cells transfected with miR-384 inhibitor. To further confirm it, wild and mutant plasmids of luciferase reporter vectors were constructed, CDC42-wt or CDC42-mut, and transfected into U251 cells with miR-384 mimics or miR-NC. Luciferase activity of CDC42-wt with transfection of miR-384 mimics group was remarkably reduced compared with that of CDC42-wt with miR-NC, which was consistent with previous outcomes. However, there was no significant difference between luciferase activity of CDC42-mut with transfection of miR-384 mimics group and that of CDC42-mut with transfection of miR-NC, which reversely demonstrated that miR-384 plays its role in suppression of glioma by targeting at CDC42.
Conclusion
To sum up, the present study demonstrated the inhibitory role of miR-384 on proliferation, migration and invasion of glioma via downregulating expression CDC42. Our results provided a little contribution on elucidating the molecular mechanism of proliferation, migration and invasion of glioma cell and a new target for therapeutic strategy against glioma.
Acknowledgment
The authors would like to thank the staff at Department of Neurosurgery, Affiliated Hospital of Jining Medical University.
Disclosure
The authors report no conflicts of interest in this work.
References
DeWitt JC, Jordan JT, Frosch MP, et al. Cost-effectiveness of IDH testing in diffuse gliomas according to the 2016 WHO classification of tumors of the central nervous system recommendations. Neuro Oncol. 2017;19(12):1640–1650. | ||
Ricard D, Idbaih A, Ducray F, Lahutte M, Hoang-Xuan K, Delattre JY. Primary brain tumours in adults. Lancet. 2012;379(9830):1984–1996. | ||
Hata A, Lieberman J. Dysregulation of microRNA biogenesis and gene silencing in cancer. Sci Signal. 2015;8(368):re3. | ||
Kim M, Kasinski AL, Slack FJ. MicroRNA therapeutics in preclinical cancer models. Lancet Oncol. 2011;12(4):319–321. | ||
Zheng HB, Zheng XG, Liu BP. miRNA-101 inhibits ovarian cancer cells proliferation and invasion by down-regulating expression of SOCS-2. Int J Clin Exp Med. 2015;8(11):20263–20270. eCollection 2015. | ||
Kouri FM, Ritner C, Stegh AH. miRNA-182 and the regulation of the glioblastoma phenotype – toward miRNA-based precision therapeutics. Cell Cycle. 2015;14(24):3794–3800. | ||
Wu F, Li J, Guo N, Wang XH, Liao YQ. MiRNA-27a promotes the proliferation and invasion of human gastric cancer MGC803 cells by targeting SFRP1 via Wnt/β-catenin signaling pathway. Am J Cancer Res. 2017;7(3):405–416. eCollection 2017. | ||
Fu Z, Li G, Li Z, et al. Endogenous miRNA Sponge LincRNA-ROR promotes proliferation, invasion and stem cell-like phenotype of pancreatic cancer cells. Cell Death Discov. 2017;3:17004. | ||
Wang G, Pan J, Zhang L, Wei Y, Wang C. Long non-coding RNA CRNDE sponges miR-384 to promote proliferation and metastasis of pancreatic cancer cells through upregulating IRS1. Cell Prolif. 2017:50(6). | ||
Sun H, He L, Ma L, et al. LncRNA CRNDE promotes cell proliferation, invasion and migration by competitively binding miR-384 in papillary thyroid cancer. Oncotarget. 2017;8(66):110552–110565. | ||
Bai PS, Xia N, Sun H, Kong Y. Pleiotrophin, a target of miR-384, promotes proliferation, metastasis and lipogenesis in HBV-related hepatocellular carcinoma. J Cell Mol Med. 2017;21(11):3023–3043. | ||
Wang YX, Chen YR, Liu SS, et al. MiR-384 inhibits human colorectal cancer metastasis by targeting KRAS and CDC42. Oncotarget. 2016;7(51):84826–84838. | ||
Mezzacappa C, Komiya Y, Habas R. Activation and function of small GTPases Rho, Rac, and Cdc42 during gastrulation. Methods Mol Biol. 2012;839:119–131. | ||
Sinha S, Yang W. Cellular signaling for activation of Rho GTPase Cdc42. Cell Signal. 2008;20(11):1927–1934. | ||
Fidyk N, Wang JB, Cerione RA. Influencing cellular transformation by modulating the rates of GTP hydrolysis by Cdc42. Biochemistry. 2006;45(25):7750–7762. | ||
Vanni C, Ottaviano C, Guo F, et al. Constitutively active Cdc42 mutant confers growth disadvantage in cell transformation. Cell Cycle. 2005;4(11):1675–1682. | ||
Lin R, Bagrodia S, Cerione R, Manor D. A novel Cdc42Hs mutant induces cellular transformation. Curr Biol. 1997;7(10):794–797. | ||
Wang JB, Erickson JW, Fuji R, et al. Targeting mitochondrial glutaminase activity inhibits oncogenic transformation. Cancer Cell. 2010;18(3):207–219. | ||
Fortin SP, Ennis MJ, Schumacher CA, et al. Cdc42 and the guanine nucleotide exchange factors Ect2 and trio mediate Fn14-induced migration and invasion of glioblastoma cells. Mol Cancer Res. 2012;10(7):958–968. | ||
Okura H, Golbourn BJ, Shahzad U, et al. A role for activated Cdc42 in glioblastoma multiforme invasion. Oncotarget. 2016;7(35):56958–56975. | ||
Reyes SB, Narayanan AS, Lee HS, et al. αvβ8 integrin interacts with RhoGDI1 to regulate Rac1 and Cdc42 activation and drive glioblastoma cell invasion. Mol Biol Cell. 2013;24(4):474–482. | ||
Tang H, Wang Z, Liu X, et al. LRRC4 inhibits glioma cell growth and invasion through a miR-185-dependent pathway. Curr Cancer Drug Targets. 2012;12(8):1032–1042. | ||
DeWitt JC, Mock A, Louis DN. The 2016 WHO classification of central nervous system tumors: what neurologists need to know. Curr Opin Neurol. 2017;30(6):643–649. | ||
Livak KJ, Schmittgen TD. Analysis of relative gene expression data using real-time quantitative PCR and the 2(-Delta Delta C(T)) Method. Methods. 2001;25(4):402–408. | ||
Chen CH, Statt S, Chiu CL, et al. Targeting myristoylated alanine-rich C kinase substrate phosphorylation site domain in lung cancer. Mechanisms and therapeutic implications. Am J Respir Crit Care Med. 2014;190(10):1127–1138. | ||
Cheng Z, Liu F, Wang G, Li Y, Zhang H, Li F. miR-133 is a key negative regulator of CDC42-PAK pathway in gastric cancer. Cell Signal. 2014;26(12):2667–2673. | ||
Fu MG, Li S, Yu TT, et al. Differential expression of miR-195 in esophageal squamous cell carcinoma and miR-195 expression inhibits tumor cell proliferation and invasion by targeting of Cdc42. FEBS Lett. 2013;587(21):3471–3479. | ||
Nandy SB, Orozco A, Lopez-Valdez R, et al. Glucose insult elicits hyperactivation of cancer stem cells through miR-424-cdc42-prdm14 signalling axis. Br J Cancer. 2017;117(11):1665–1675. | ||
Shi C, Ren L, Sun C, et al. miR-29a/b/c function as invasion suppressors for gliomas by targeting CDC42 and predict the prognosis of patients. Br J Cancer. 2017;117(7):1036–1047. | ||
van der Sanden B, Ratel D, Berger F, Wion D. Glioma recurrence following surgery: peritumoral or perilesional? Front Neurol. 2016;7:52. | ||
A Mirza F, Shamim MS. Extent of resection and timing of surgery in adult low grade glioma. J Pak Med Assoc. 2017;67(6):959–961. |
 © 2018 The Author(s). This work is published and licensed by Dove Medical Press Limited. The full terms of this license are available at https://www.dovepress.com/terms.php and incorporate the Creative Commons Attribution - Non Commercial (unported, v3.0) License.
By accessing the work you hereby accept the Terms. Non-commercial uses of the work are permitted without any further permission from Dove Medical Press Limited, provided the work is properly attributed. For permission for commercial use of this work, please see paragraphs 4.2 and 5 of our Terms.
© 2018 The Author(s). This work is published and licensed by Dove Medical Press Limited. The full terms of this license are available at https://www.dovepress.com/terms.php and incorporate the Creative Commons Attribution - Non Commercial (unported, v3.0) License.
By accessing the work you hereby accept the Terms. Non-commercial uses of the work are permitted without any further permission from Dove Medical Press Limited, provided the work is properly attributed. For permission for commercial use of this work, please see paragraphs 4.2 and 5 of our Terms.