Back to Journals » Cancer Management and Research » Volume 10
MicroRNA-365 suppresses cell growth and invasion in esophageal squamous cell carcinoma by modulating phosphoserine aminotransferase 1
Authors Sun C, Zhang X, Chen Y, Jia Q, Yang J, Shu Y
Received 23 November 2017
Accepted for publication 22 January 2018
Published 15 October 2018 Volume 2018:10 Pages 4581—4590
DOI https://doi.org/10.2147/CMAR.S157858
Checked for plagiarism Yes
Review by Single anonymous peer review
Peer reviewer comments 2
Editor who approved publication: Dr Kenan Onel
Changjiang Sun,1 Xizhi Zhang,1 Yong Chen,1 Qingqing Jia,1 Jianqi Yang,1 Yusheng Shu2
1Department of Oncology, Subei People’s Hospital of Jiangsu Province, Yangzhou, Jiangsu, China; 2Department of Thoracic and Cardiovascular Surgery, Subei People’s Hospital of Jiangsu Province, Yangzhou, Jiangsu, China
Background: A number of studies have indicated that expression of miRNA-365 (miR-365) is suppressed in various cancers, suggesting its cancer-suppressive role. In the present investigation, we evaluated the regulation and character of miR-365 in human esophageal squamous cell carcinoma (ESCC).
Patients and methods: The tumor tissues and adjacent nontumor tissue samples were collected from 30 patients having ESCC, and the expression levels of miR-365 were studied by quantitative real-time polymerase chain reaction (PCR). MTT and cell invasion by Matrigel assay were done to study the effect of miR-365 on proliferation and metastasis of ESCC cells. An in vivo tumor model was generated by inoculating ESCC cells subcutaneously into BALB nude mice. A study of various biomarkers such as quantitative polymerase chain reaction (qPCR), luciferase activity assay, and Western blot was done to confirm the targets of miR-365.
Results: In tumor tissues, a significant downregulation of miR-365 was observed versus the nontumor adjacent tissues and ESCC cells versus the selected esophageal endothelial cells. It was observed that higher expression levels of miR-365 inhibited the cell invasion, colony formation, growth in esophageal cancer cell lines in vitro, and tumor development in vivo. The study of biomarkers suggests involvement of phosphoserine aminotransferase 1 (PSAT1) as a favorable target for miR-365, and its abnormal expression inverted the miR-365-arbitrated suppression of invasion, viability, and epithelial–mesenchymal transition in esophageal cancer cells. A negative correlation existed with expression of miR-365 and PSAT1 in human esophageal cancer tissue samples.
Conclusion: The study established that miR-365 exhibits tumor-suppressive action via regulating the levels of PSAT1 and leads to invasion and progressiveness of esophageal cancer.
Keywords: esophageal cancer, miR-365, PSAT1, tumor
Introduction
Esophageal squamous cell carcinoma (ESCC), which also includes esophagus adenocarcinoma, ranks sixth among all types of cancers causing mortality and eighth in the list of common cancers worldwide.1,2 ESCC is reported to be the most happening cancer (70%) among all the reported cases of esophageal cancer worldwide, and in China, the figures are even more alarming as the occurrence of ESCC is at least 30 times more compared with that in USA.3 ESCC originates from the esophagus, mostly from the epithelial cell linings. The etiology mainly associated with the occurrence of ESCC includes smoking, poor diet plan, alcoholism, and hot drinks.1 The high mortality rate in ESCC is mainly due its late diagnosis and its aggressive nature.
miRNAs are noncoding RNA molecules having single strand (23 nucleotides), and these molecules can bring about regulation of gene expression via binding to the 3′-untranslated regions (UTRs) of target genes at the posttranscriptional level.4,5 Recently, the human genome has been reported to show presence of a large number of miRNAs (>1000) that regulate ~30% of all the discovered genes.6 A number of studies have indicated that deviant levels of miRNAs are linked with disorders such as cancer; based on the type of mRNAs they can act as tumor suppressors.7–11 The miRNA-365 (miR-365) is confirmed for its tumor-suppressive effect in a number of cancers such as cutaneous squamous cell carcinoma,12 lung,13 breast,14 and hepatic carcinoma.15 ESCC has been reported for its aberrant expression of miRNAs identified using quantitative reverse transcription polymerase chain reaction (qRT-PCR);16,17 however, studies suggesting the correlation between expression of miR-365 and ESCC along with specific mechanisms of miR-365 in development of esophageal cancer have not been reported yet.
In cancer, the cells tend to grow rapidly and hence alter metabolic pathways, and to meet these cell requirements, metabolic pathways affecting the production of glycine and serine play an important role.18 Recently, a number of tumorigenesis enzymes have been identified, which are involved in biologic reactions facilitating to maintain the redox status and the biosynthesis of cellular components.19,20 Among them, phosphoserine aminotransferase 1 (PSAT1) and phosphoglycerate dehydrogenase (PHGDH) are the two enzymes responsible for the catalysis of serine. These enzymes are found to be upregulated in a number of cancers and are reported to be responsible by diverting the glucose flux for metabolism of serine.20 However, the regulatory mechanism behind the expression of these enzymes and their role in cell proliferation are still not clearly understood.
In the present research, we first established that expression of miR-365 was significantly downregulated in ESCC cell lines and tissues. Our study further confirmed the tumor-suppressive activity of miR-365 in ESCC via the PSAT1 pathway. The outcomes of our study confirmed a new therapeutic target for ESCC.
Patients and methods
Human esophageal tissue specimens
A total of 30 patients diagnosed with esophageal carcinoma in 2016 and who underwent surgery for esophageal resection before submitting to chemotherapy at Subei People’s Hospital of Jiangsu Province were selected for this study. From these patients, tissue specimens of ESCC and the adjacent area (normal tissue) were collected after surgery under the guidelines from an experienced pathologist. Before storing in liquid nitrogen, the tissues were dissected to remove any connective tissue. The tissue samples were stored in the tissue bank of Subei People’s Hospital of Jiangsu Province, and on receiving, the samples were coded in accordance with the guidelines from the ethical committee. All the protocols were in accordance with the local human ethical committee of Subei People’s Hospital of Jiangsu Province followed by the approval from hospital board (sanctioned reference number HEC/SPHJ/ESO/14B-16/17). Written informed consent was obtained from all patients.
Cell lines and culture conditions for in vitro studies
The human esophageal cancer cell lines were procured from Cell Collection Center, Shanghai (Chinese Academy of Science-China). Three ESCC cell lines (EC109, EC1, and EC9706), HEEC, and HEK293T cells for transfection were used in the present study and were obtained from Chinese Academy of Medical Science (Shanghai, China); the cells were maintained in DMEM supplemented with 10% fetal bovine serum (FBS), 100 μg/mL streptomycin, and 100 units/mL penicillin G at 37°C and 5% CO2. The cells were grown in EpiCM-2 along with EpiCGS-2 (Innoprot, Derio, Spain).
Extraction of RNA and qRT-PCR studies
Total miRNA was isolated from the cells using an miRNeasy kit (Qiagen GmbH, Hilden, Germany) in accordance with the manufacturer’s protocol; next, an miScriptRT kit was used to convert it into cDNA. RNase-free water was used to elute RNA, followed by recording the absorbance at a wavelength of 280 nm, and then used for reverse transcription. TaqMan assay was done by using TaqMan Human miRNA Array Panel (Thermo Fisher Scientific, Waltham, MA, USA) in accordance with the manufacturer’s instructions, and the expression was normalized using endogenous U6. The miR-365 mimics and negative control RNA were from GenePharma (Shanghai, China). For transcription of PSAT1, iScript cDNA synthesis kit (Bio-Rad Laboratories Inc., Hercules, CA, USA) was used as per the manufacturer’s instructions for synthesizing cDNA. Real-time qPCR was done using an ABI 7,500 Real-Time PCR System (Thermo Fisher Scentific). The stem-loop RT primer, qRT-PCR primers, and the internal control U6 primers were used (Table 1). U6 was opted as internal control for relative expression of miR-365. Each trial was performed three times.
  | Table 1 The sequence of primers Abbreviations: qRT-PCR, quantitative reverse transcription polymerase chain reaction; RT, reverse transcription. |
Construction of plasmid and transduction of lentiviral and gene transfection
To construct the miR-365, lentiviral gene transfer vector carrying hsa-miR-365, amplification of fragment-encoding sequence for pre-miR-365 was done by PCR followed by cloning into the sites of pGCsil-green fluorescence protein (GFP) vector (Shanghai Genechem Co., Ltd, Shanghai, China) to get the lentivirus-mediated miR-365 vector (LV-miR-365). To generate the PSAT1 overexpression plasmid, a pcDNA3 vector was used. The amplification of PSAT1 cDNA was done by PCR using cDNA extracted from HL60 cell lines. After amplification, the PSAT1 cDNA gene was cloned into the vector.
The 3′-UTR fragment of PSAT1 having the recognition sequence of miR-365 was subjected to amplification using PCR using the required primers. After amplification, subcloning was done into the vector (psiCHECK-2; Promega Corporation, Fitchburg, WI, USA) to construct PSAT1 3′-UTR clone using Phusion Site-Directed Mutagenesis Kit (Thermo Fisher Scientific). The pCDH cDNA was transfected in the PSAT1 packaging system for generating virus particles using Lipofectamine 2000 reagent (Thermo Fisher Scientific) into HEK293T cells as per the manufacturer’s instruction. The cells, after growing to the log phase, were transfected with pGCsil-GFP vector for 24 h. Mass screening of stable cells was done after looking into the levels of GFP mediated by lentivirus vector post 72 h of transfection using a flow cytometer (BD Biosciences, San Jose, CA, USA).
For carrying out gene transfection, the ESCC cells (5×105) were transfected in 2 mL culture medium with 100 pmol control vector in a serum-free medium with Lipofectamine reagent. The culture medium was changed after 6 h, and the cells were maintained in an incubator at 37°C in 5% CO2 for 48 h.
3′-UTR dual-luciferase reporter assay and the human embryonic kidney cells 293
To build the luciferase reporter vector, the binding sites for miR-365 in the PSAT1 and the PSAT1 3-UTR fragment were amplified by PCR using the reverse primers. The amplified protein was then inserted into the luciferase gene psiCHECK2 vector, named as PSAT1 3′-UTR-wild. Phusion Site-Directed Mutagenesis Kit (Thermo Fisher Scientific) was used for site-directed mutagenesis of miR-365 of the target site in PSAT1 3′-UTR, named as PSAT1 3′-UTR-mut. The transfection was done with miR-365 mimics and with either PSAT1 3′-UTR-mut or wild. The luciferase assay was done 48 h post-transfection using Luciferase Reporter Gene Detection Kit (Sigma-Aldrich Co., St Louis, MO, USA).
Cell invasion assay
For the assay, a Matrigel-coated (1:5 serum-free RPMI medium-1640) membrane filter (8 μm) was used. The membrane filter was subjected to sterilization and air-drying, and then 20 μL of medium was added into the bottom chamber of corning Transwell® plates. The transfected cells (1.5×106 cells/mL) were resuspended in serum-free RPMI medium and were subjected to seeding in the upper space of Transwell® plates followed by incubation for 24 h at 37°C. The cells on the bottom of the filter were fixed in methanol at −20°C for 15 min followed by washing with PBS and later staining of crystal violet, and the cells were counted under a microscope.
Cell viability studies (MTT assay)
The in vitro cell viability of EC109 and EC9706 cells was evaluated by MTT assay using Vybrant® MTT Cell Proliferation Assay Kit (Thermo Fisher Scientific) following the manufacturer’s protocol. The procedure involved replacement of medium with fresh medium containing MTT (0.5 mg/mL) at defined time intervals indicated by the manufacturer. The cells were then subjected to incubation for 1 h at 37°C resolved by the provided reagent followed by measurement of absorbance at 570 nm.
Colony formation assay
The EC109 and EC9706 cells were trypsinized and suspended and cultured in culture plates (60 mm diameter) along with 10% FBS for 2 weeks. The staining and fixing of developed colonies were done using 0.5% crystal violet solution for 20 min followed by washing (three times). Clusters with cell count >50 were counted as colonies. Microscopic examination and counting of colonies were done from 10 randomly selected viewed fields, and the mean representing the 95% confidence region was attained.
Tumorigenicity studies in BALB/c Scid mice (in vivo studies)
All the animal experiments were performed in accordance with the approved animal protocols and guidelines established by the Institutional Ethical Committee of Subei People’s Hospital of Jiangsu Province with approval number SPH/AIC/00241M. The experiments followed the guidelines provided by the committee, which were in accordance with the draft of “Animal protection law of the People’s Republic of China-2009” for experimental animals. The experimental BALB/C nude mice used were obtained from Beijing University Experimental Animal Center (China). The animals were housed in a pathogen-free condition. For inducing tumor, the mice were injected with 200 μL of serum-free medium having ~1×106 cells/mL subcutaneously into the upper flank. The growth and volumes of esophageal tumors were measured with calipers once every 7 days. After killing the animals (5 weeks), the tumor weights were examined.
Expression of proteins by Western blotting
After receiving the treatment, the cells were subjected to lysis using RIPA buffer (Cell Signaling Tech, Inc, Danvers, MA, USA), and the extracts were submitted to sodium dodecyl sulfate–polyacrylamide gel electrophoresis followed by transfer to nitrocellulose membranes and blotting. The membranes were blocked using a mixture of 5% skim milk and 0.1% Tween 20 in Tris-buffered saline. The membranes were incubated with Iry antibodies at 4°C for 12 h. PSAT1, p-GSK-3 β, Snail, E-cadherin, GSK-3β, Vimentin (Thermo Fisher Scientific), and the anti-GAPDH antibody were used as internal controls (Thermo Fisher Scientific). The blots were visualized using enhanced chemiluminescence after incubating with secondary antibodies for 2 h.
Statistical data analysis and significance
All the data reported were average±SD (n=3). One-way analysis of variance or t-test (paired) was used for establishing differences. The values of p <0.05 were regarded as statistically significant. The expression of relative proteins by Western blotting was done, using GAPDH as loading control.
Results
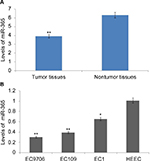
Levels of miR-365 are decreased in EC1, EC109, and EC9706 cell lines and tissues
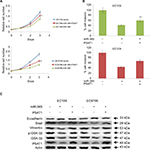
The esophageal cancerous tissues and the proximate noncancerous tissues in the same subjects were obtained from esophageal cancer patients (30 numbers), and the tissues were subjected to qRT-PCR for comparing the expressions of miR-365. The outcome of the experiment showed that the expression of miR-365 was significantly (P<0.01) toward lower side in tumor tissues recovered from patients compared to nontumor adjacent tissues (Figure 1A). The miR-365 levels in ESCC cells, that is, EC109, EC1, and EC9706, were similar to those observed in tissue samples, and the levels of miR-365 were on the higher side when compared with the normal HEEC cell lines. The expression levels were toward the higher side in EC109 and the lowest in EC9706 ESCC cell lines (Figure 1B); hence, EC109 and EC9706 were selected for experiments in the study.
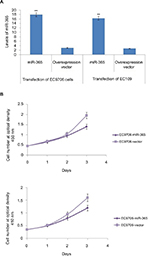
MiR-365 suppresses the growth of ESCC cells (in vivo and in vitro)
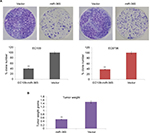
To study the effect of miR-365 on esophageal cancer in vivo and in vitro, the EC9706 and EC109 cells were transfected with overexpression vector of miR-365, and the cell viability and tumor growth were evaluated. qRT-PCR was utilized for confirming the overexpression levels of miR-365 in EC109 and EC9706 cell lines (Figure 2A). The overexpression of miR-365 significantly (P<0.01) suppressed cell viability as reported by MTT assay and colony formation by colony-forming assay in both the esophageal cancer cells (Figures 2B and 3A). Tumor growth was evaluated in the xenografts mouse model, and the tumor weight reduced significantly (P<0.01) (Figure 3B) in mice receiving injection of EC9706 cells overexpressed for miR-365 compared with those receiving injection of vector control cells.
Inhibitory action of miR-365 on cell invasion in ESCC cell lines
Cell invasion or Matrigel invasion assay was done to analyze the role of miR-365 vector transfection in esophageal cell lines. The results demonstrated that miR-365 overexpression vector suppressed the invasion of selected EC9706 and EC109 cell lines significantly (P<0.01) by ~64% compared to control (Figure 4A and B).
Identification of PSAT1 as a favorable target site of miR-365
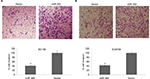
To identify the molecular mechanism behind miR-365, the experiments using prediction tools were designed to examine the potential targets. A prediction algorithm of Targetscan was used to identify the conserved algorithm domain within 3′-UTR of PSAT1 having two bonding sites for miR-365. To establish this prevision, luciferase activity was evaluated to validate PSAT1 as a target for miR-365. The constructed luciferase reporter plasmids built by cloning the mutant 3′-UTR of PSAT1-expressing mutations in the miR-365 overexpression vector and the miR-365 seed region were used to co-transfect the HEK293 cells. On assessing the luciferase activity, it was found that the overexpression of miR-365 did not suppress the reporter gene activity for mutations of PSAT 3′-UTR on two binding sites but significantly suppressed the activity of the wild-type PSAT1-UTR-MUT1 or MUT2 (P<0.01) (Figure 5A). The abnormal expression of miR-365 decreased the mRNA expression of PSAT1 significantly compared to the loading control, that is, GAPDH in both the ESCC cell lines (Figure 5B), confirming PSAT1 as a favorable target for miR-365.
The outcomes of miR-365 on tumor invasion were analyzed by expression of epithelial markers such as E-cadherin, Snail, Vimentin, the Ser/Thr kinase GSK-3β, and EMT, that is, epithelial–mesenchymal transition markers. Reports suggest a correlation between the transcription factors Snail and EMT; Snail modulates the levels of E-cadherin, whereas downregulation of E-cadherin stimulates the activity of EMT. Reports have established that Ser/Thr kinase family protein GSK-3β is deactivated as a result of phosphorylation at Ser9. GSK-3β is found to regulate Snail activity; also, the epithelial phenol-typing is maintained by suppressing Snail, and hence, deactivation of GSK-3β elevates EMT.21 The results of Western blot as depicted in Figure 5C suggested that overexpression of miR-365 increased the expression levels of E-cadherin, suppressed the levels of Snail as well as Vimentin, and decreased the levels of total and associated phosphorylated GSK-3β followed by decrease in the levels of PSAT1. The results of overall protein expression suggested that miR-365 could inhibit EMT associated with downregulation of PSAT1 in cancerous ESCC cell lines.
Involvement of PSAT1 in miR-365-mediated suppression of cell proliferation and invasion in ESCC cell lines
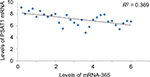
To further confirm the role of miR-365-mediated suppression of PSAT1, in correlation to proliferation and invasion of ESCC cell lines. The study showed that the expression of PSAT1 was ectopic, which in turn partly reclaimed the suppression of cell invasion and proliferation caused by overexpression of miR-365 in ESCC cell lines (Figure 6A and B). The study also showed that the ectopic expression of PSAT1 resulted in partial restoration of the E-cadherin and Snail levels, which were partially suppressed by miR-365, confirming the role of PSAT1 in reversing the consequences of miR-365 on EMT in selected esophageal cell lines (Figure 6C). A negative correlation was established between levels of mRNA of PSAT1 and expression of miR-365 by qRT-PCR studies involving miR-365 and levels of PSAT1 in cancerous tissue samples from patients reported with esophageal carcinoma (P<0.01, R2=0.369). The study established that miR-365 is involved in the regulation and expression of PSAT1 in ESCC (Figure 7).
Discussion
Here, we studied the role and extent of expression of miR-365 in ESCC and also the mechanism involved. We demonstrated that ESCC (cell lines and ESCC-affected human tissue) is associated with the downregulation of miR-365, and its overexpression halts its progression. Our findings suggested that the overexpression of miR-365 resulted in inhibition of colony formation, cell viability, and cell invasion in cancerous esophageal cells and also inhibited the growth of tumor in xenografts mice. The study discovered PSAT1 as a favorable target of miR-365 in esophageal tumor tissues. The findings suggested that miR-365 can behave as a tumor suppressor by regulating the levels of PSAT1.
Esophageal cancer is colligated with both inadequate prognosis and survival.22,23 Advanced-stage diagnosis has attributed to the poor survival of ESCC patients, thus creating the necessity to discover specific biologic markers for its early-stage detection. A number of reports have worked on discovering miRNAs for distinguishing ESCC from normal tissues using microarray analysis.
In a study involved in the microarray analysis of human miRNAs in cancerous ESCC tissues, out of 210 miRNAs, about four, that is, miR-373, let-7d, miR-340, and miR-330, were upregulated.16 A qRT-PCR analysis of ESCC tissues done by Ogawa et al identified expression of 10 miRNAs and reported six of them to be overexpressed and four downregulated;24 similarly, a study by Kano et al. reported expression of 15 miRNAs downregulated in ESCC tissues.25 Similarly, a study involved in identifying miRNAs reported 31 miRNAs in correlation with ESCC,26 among which seven were able to distinguish ESCC from the normal tissues. The study confirmed that the expression levels of miR-140, -29c, -99a, and -100 were downregulated, while those of miR-151, miR-424, and miR-25 were upregulated.26 Yan et al27 reported decreased expression of miR-340 in ESCC and found that miR-340 caused inhibition by targeting PSAT1.28
Although a number of studies have dug into the role of miRNAs in the progression of ESCC, the identified potential targets need further confirmation. Kano et al found that the three miRNAs (miR-133a, mir-133b, and miR-145) upon transfection into ESCC cells caused inhibition of cell proliferation and invasion.25 In a study related to ESCC, it was found that miR-25 inhibited cadherin 1 activity.29 Similarly, many potential targets were identified in other studies involving ESCC, which include the ERK5 pathway for miR-143, ZEB1 for miR-150, Rap1b for miR-518b, and FBXW7 for miR-223.30
In the present investigation, we established that miR-365 is downregulated in esophageal tumor tissues and ESCC cell lines, and we also evidenced that the expression of miR-365 is linked to proliferation, and colony-forming and invasion capability of cancerous esophageal cells in addition to the growth of tumor in mice. The study established that miR-365 exhibited tumor-suppressing activity in esophageal cancer by aiming the PSAT1 pathway. The outcome of experiments demonstrated that the overexpression of miR-365 resulted in the downregulation of PSAT1, and the abnormal expression of PSAT1 reversed the inhibitory activity of miR-365 on cell proliferation, invasion, and expression of EMT.
PHGDH and PSAT1 are the enzymes of serine pathway; literature suggest modifications in the levels of these two associated with progression of many cancers such as breast cancer.31 The expression of PHGDH is reported to be increased in some cases of breast cancer, and the downregulation of PSAT1 exhibited antiproliferative effect in such PHGDH-overexpressing breast cancer cells.32,33 The overexpression of PSAT1 in colon cancer is associated with chemoresistance.31 A weakened response for endocrine treatment approach in breast cancer is linked to overexpression of PSAT1.33 All the reports confirmed participation of PSAT1 in progression of cancer, which adheres to our present findings suggesting that suppression of miR-365 may result in tumorigenesis in esophageal cancer by upregulation of PSAT1. In future, experiments focusing on the correlation between PSAT1 and miR-365 may further strengthen their roles in treating ESCC.
Conclusion
The outcomes of the present study confirmed tumor suppressor effect of miR-365 in ESCC by modulating the expression levels of PSAT1. The study confirmed that the effect of miR-365 on cell invasion and proliferation gets reversed after abnormal expression of PSAT1. Hence, this study confirmed that miR-365 exerts antitumor effect via PSAT1 pathway and hence suggests a novel biomarker in the development and progression of ESCC.
Acknowledgments
We express our thankful concerns to the staff and management of Subei People’s Hospital of Jiangsu Province, Yangzhou, for providing necessary facilities. We also thank Wei Wang from College of Medicine, Fudan University for their kind help during experimental work.
Disclosure
The authors report no conflicts of interest in this work.
References
Arnold M, Soerjomataram I, Ferlay J, Forman D. Global incidence of oesophageal cancer by histological subtype in 2012. Gut. 2015;64:381–387. | ||
Lagergren J. Oesophageal cancer in 2014: advances in curatively intended treatment. Nat Rev Gastroenterol Hepatol. 2015;12:74–75. | ||
Zhang Y. Epidemiology of esophageal cancer. World J Gastroenterol. 2013;19:5598–5606. | ||
Bartel DP. MicroRNAs: target recognition and regulatory functions. Cell. 2009;136:215–233. | ||
He L, Hannon GJ. MicroRNAs: small RNAs with a big role in gene regulation. Nat Rev Genet. 2004;5:522–531. | ||
Nana-Sinkam SP, Croce CM. Clinical applications for microRNAs in cancer. Clin Pharmacol Ther. 2013;93:98–104. | ||
Osman A. MicroRNAs in health and disease – basic science and clinical applications. Clin Lab. 2012;58:393–402. | ||
Esquela-Kerscher A, Slack FJ. Oncomirs - microRNAs with a role in cancer. Nat Rev Cancer. 2006;6:259–269. | ||
Wang QX, Zhu YQ, Zhang H, Xiao J. Altered MiRNA expression in gastric cancer: a systematic review and meta-analysis. Cell Physiol Biochem. 2015;35:933–944. | ||
Li W, Jiang G, Zhou J, et al. Down-regulation of miR-140 induces EMT and promotes invasion by targeting Slug in esophageal cancer. Cell Physiol Biochem. 2014;34:1466–1476. | ||
Zhu Y, Xia Y, Niu H, Chen Y. MiR-16 induced the suppression of cell apoptosis while promote proliferation in esophageal squamous cell carcinoma. Cell Physiol Biochem. 2014;33:1340–1348. | ||
Zhou L, Gao R, Wang Y, Zhou M, Ding Z. Loss of BAX by miR-365 promotes cutaneous squamous cell carcinoma progression by suppressing apoptosis. Int J Mol Sci. 2017;18(6):1157. | ||
Qi J, Rice SJ, Salzberg AC, et al. MiR-365 regulates lung cancer and developmental gene thyroid transcription factor 1. Cell Cycle. 2012;11(1):177–186. | ||
Hong B, Li H, Zhang M, et al. p38 MAPK inhibits breast cancer metastasis through regulation of stromal expansion. Int J Cancer. 2015;136(1):34–43. | ||
Chen Z, Huang Z, Ye Q, et al. Prognostic significance and anti-proliferation effect of microRNA-365 in hepatocellular carcinoma. Int J Clin Exp Pathol. 2015;8(2):1705–1711. | ||
Lee KH, Goan YG, Hsiao M, et al. MicroRNA-373 (miR-373) post-transcriptionally regulates large tumor suppressor, homolog 2 (LATS2) and stimulates proliferation in human esophageal cancer. Exp Cell Res. 2009;315:2529–2538. | ||
Zhang M, Yang Q, Zhang L, et al. MiR-302b is a potential molecular marker of esophageal squamous cell carcinoma and functions as a tumor suppressor by targeting ErbB4. J Exp Clin Cancer Res. 2014;33:10. | ||
Ma L, Tao Y, Duran A, et al. Control of nutrient stress-induced metabolic reprogramming by PKCzeta in tumorigenesis. Cell. 2013;152:599–611. | ||
Chen J, Gammon MD, Chan W, et al. One-carbon metabolism, MTHFR polymorphisms, and risk of breast cancer. Cancer Res. 2005;65:1606–1614. | ||
Locasale JW, Grassian AR, Melman T, et al. Phosphoglycerate dehydrogenase diverts glycolytic flux and contributes to oncogenesis. Nat Genet. 2011;43:869–874. | ||
Kim JY, Kim YM, Yang CH, Cho SK, Lee JW, Cho M. Functional regulation of Slug/Snail2 is dependent on GSK-3beta-mediated phosphorylation. FEBS J. 2012;279:2929–2939. | ||
Jemal A, Siegel R, Xu J, Ward E. Cancer statistics, 2010. CA Cancer J Clin. 2010;60:277–300. | ||
Li YJ, Zhou JH, Du XX, et al. Dihydroartemisinin accentuates the anti-tumor effects of photodynamic therapy via inactivation of NF-κB in Eca109 and Ec9706 esophageal cancer cells. Cell Physiol Biochem. 2014;33:1527–1536. | ||
Ogawa R, Ishiguro H, Kuwabara Y, et al. Expression profiling of micro-RNAs in human esophageal squamous cell carcinoma using RT-PCR. Med Mol Morphol. 2009;42:102–109. | ||
Kano M, Seki N, Kikkawa N, et al. MiR-145, miR-133a and miR-133b: tumor-suppressive miRNAs target FSCN1 in esophageal squamous cell carcinoma. Int J Cancer. 2010;127:2804–2814. | ||
Guo Y, Chen Z, Zhang L, et al. Distinctive microRNA profiles relating to patient survival in esophageal squamous cell carcinoma. Cancer Res. 2008;68:26–33. | ||
Yan S, Jiang H, Fang H, et al. MicroRNA-340 inhibits esophageal cancer cell growth and invasion by targeting phosphoserine aminotransferase 1. Cell Physiol Biochem. 2015;37:375–386. | ||
Sun Y, Yu X, Bai Q. miR-204 inhibits invasion and epithelial-mesenchymal transition by targeting FOXM1 in esophageal cancer. Int J Clin Exp Pathol. 2015;8:12775-12783. | ||
Xu X, Chen Z, Zhao X, et al. MicroRNA-25 promotes cell migration and invasion in esophageal squamous cell carcinoma. Biochem Biophys Res Commun. 2012;421:640–645. | ||
Sakai NS, Samia-Aly E, Barbera M, Fitzgerald RC. A review of the current understanding and clinical utility of miRNAs in esophageal cancer. Semin Cancer Biol. 2013;23:512–521. | ||
Vie N, Copois V, Bascoul-Mollevi C, et al. Overexpression of phosphoserine aminotransferase PSAT1 stimulates cell growth and increases chemoresistance of colon cancer cells. Mol Cancer. 2008;7:14. | ||
Pollari S, Kakonen SM, Edgren H, et al. Enhanced serine production by bone metastatic breast cancer cells stimulates osteoclastogenesis. Breast Cancer Res Treat. 2011;125:421–430. | ||
Martens JW, Nimmrich I, Koenig T, et al. Association of DNA methylation of phosphoserine aminotransferase with response to endocrine therapy in patients with recurrent breast cancer. Cancer Res. 2005;65:4101–4117. |
 © 2018 The Author(s). This work is published and licensed by Dove Medical Press Limited. The full terms of this license are available at https://www.dovepress.com/terms.php and incorporate the Creative Commons Attribution - Non Commercial (unported, v3.0) License.
By accessing the work you hereby accept the Terms. Non-commercial uses of the work are permitted without any further permission from Dove Medical Press Limited, provided the work is properly attributed. For permission for commercial use of this work, please see paragraphs 4.2 and 5 of our Terms.
© 2018 The Author(s). This work is published and licensed by Dove Medical Press Limited. The full terms of this license are available at https://www.dovepress.com/terms.php and incorporate the Creative Commons Attribution - Non Commercial (unported, v3.0) License.
By accessing the work you hereby accept the Terms. Non-commercial uses of the work are permitted without any further permission from Dove Medical Press Limited, provided the work is properly attributed. For permission for commercial use of this work, please see paragraphs 4.2 and 5 of our Terms.