Back to Journals » OncoTargets and Therapy » Volume 11
MicroRNA-302a-3p suppresses hepatocellular carcinoma progression by inhibiting proliferation and invasion
Authors Ye Y, Song Y, Zhuang J, Wang G, Ni J, Zhang S, Xia W
Received 3 March 2018
Accepted for publication 23 May 2018
Published 10 December 2018 Volume 2018:11 Pages 8175—8184
DOI https://doi.org/10.2147/OTT.S167162
Checked for plagiarism Yes
Review by Single anonymous peer review
Peer reviewer comments 2
Editor who approved publication: Prof. Dr. Geoffrey Pietersz
Ying Ye,1,* Yanan Song,1,* Juhua Zhuang,1 Guoyu Wang,1 Jing Ni,1 Suiliang Zhang,2 Wei Xia1
1Department of Nuclear Medicine, The Seventh People’s Hospital, Shanghai University of Traditional Chinese Medicine, Shanghai, People’s Republic of China; 2Oncology Department, The Seventh People’s Hospital, Shanghai University of Traditional Chinese Medicine, Shanghai, People’s Republic of China
*These authors contributed equally to this work
Background: Involvement of microRNAs in tumor development and their potential as prognostic biomarkers had been well acknowledged. However, the expression, clinical significance, and functional mechanisms of microRNA (miR)-302a-3p in hepatocellular carcinoma (HCC) have not been reported.
Patients and methods: Real-time quantitative polymerase chain reaction was used to evaluate the expression of miR-302a-3p in 111 HCC tissues and adjacent normal liver tissues. Its association with clinicopathological characteristics was analyzed by the chi-square test. The Kaplan–Meier univariate survival analysis and multivariate Cox regression analysis were used to identify the clinical significance of miR-302a-3p in the overall survival (OS) of HCC patients. Transfection of miR-302a-3p mimics into HepG2 and Huh7 HCC cell lines was conducted to reveal its underlying mechanism in regulating HCC progression.
Results: miR-302a-3p expression was significantly decreased in HCC tissues compared with that in paired adjacent normal liver tissues (P=0.005). miR-302a-3p expression was correlated with tumor number (P=0.003), tumor size (P<0.001), and tumor TNM stage (P=0.028). The Kaplan–Meier survival analysis showed that patients in the high miR-302a-3p expression group had a better OS than those in the low miR-302a-3p expression group (P=0.002). Multivariate analysis confirmed that miR-302a-3p expression can be used as an independent predictor for HCC prognosis (HR=0.480, 95% CI=0.249–0.894, P=0.039). Proliferation, migration, and invasion capacities were all decreased in cells transfected with miR-302a-3p mimics. Moreover, our data showed a direct effect of miR302a-3p on inhibiting the expression and signaling of PRKACB in HCC cells.
Conclusions: miR-302a-3p serves as a tumor suppressor in HCC progression by directly inhibiting tumor proliferation and invasion, and its low expression is a potential biomarker for predicting a poor prognosis of HCC patients.
Keywords: hepatocellular carcinoma, miR-302a-3p, invasion, prognosis, proliferation
Introduction
Hepatocellular carcinoma (HCC) is the most frequent tumor type of primary liver cancer, accounting for the third highest cause of cancer-related deaths.1 HCC usually occurs subsequently to liver cirrhosis caused by sustained alcohol intake or to chronic hepatitis virus (HBV, HCV, etc) infection.2 HCC is characterized by its high rate of distant metastasis and tumor recurrence, even after curative resection treatment.3 Accordingly, HCC is a highly malignant tumor type, and the 5-year overall survival (OS) is reported to be 30%–50%.4 Therefore, identifying novel biomarkers for predicting prognosis and chemotherapy treatment is of great importance. Although multiple tumor biomarkers have been reported to regulate HCC occurrence and progression, our knowledge on its underlying molecular mechanisms remains limited.
Besides conventional protein markers, recent advances in genomics and metabolomics have made great contributions to the identification of novel tumor biomarkers. miRNAs are endogenous noncoding RNAs comprising ~20 nucleotides.5 Their general function is to inhibit the protein expression by binding to the 3′-untranslated region (UTR) of targeted mRNAs.6 By modulating RNA modification and protein translation, microRNAs are considered critical in human diseases including tumor development.7 Moreover, more and more microRNAs are acknowledged as tumor biomarkers; they can help not only diagnose tumor occurrence and distinguish tumor subtype but also predict patients’ prognosis.8 The advantages of microRNAs as clinically potential markers include their stable properties and their feasibility to be detected by high-throughput strategies. For example, the miR-302 has been reported to be a prognostic biomarker in several tumor types, including glioma,9 renal carcinoma,10 and gastric cancer.11 Of note, miR-302 was recently revealed to inhibit the endothelial–mesenchymal transition of endothelial cells, thus indirectly regulating the progression of HCC according to cellular experiments.12 However, whether miR-302a-3p was aberrantly expressed in HCC tissues and its direct effect on HCC progression have not been investigated.
Here, in this study, we first explored the expression of miR-302a-3p in HCC tissues and statistically analyzed its clinical significance as a novel prognostic biomarker. Furthermore, we conducted cellular experiments by overexpressing miR-302a-3p mimics to better illustrate its tumor-promoting role in HCC development.
Patients and methods
Patients and tissues
This study was approved by the ethics committee of The Seventh People’s Hospital. A total of 111 HCC tissues and paired adjacent specimens were collected from the Department of Pathology in our hospital. The usage of tissue samples was conducted after obtaining all written informed consents from corresponding patients. All tissues were resected between 2009 and 2013 and embedded in paraffin. The inclusion criteria included a precise pathological diagnosis without any preoperative therapy, as well as the availability of follow-up data.
miRNA extraction and real-time quantitative polymerase chain reaction (RT-qPCR)
miRNAs were extracted from tissues using a miRNeasy mini kit (Qiagen NV, Venlo, the Netherlands) according to the manufacturer’s instructions. The quality and concentration of isolated RNAs were confirmed by a NanoDrop ND-1000 Spectrophotometer (Thermo Fisher Scientific). A total amount of 50 ng RNAs was reversely transcribed into cDNA using a TaqMan miRNA Reverse Transcription Kit (Thermo Fisher Scientific) according to the standard protocol. The RT-qPCR was performed using a real-time polymerase chain reaction (PCR) instrument (Thermo Fisher Scientific) using the primers as follows: miR-302a-3p forward: 5′-AATAAGTGCTTCCATGTTTTGGTGA-3′13 and GAPDH forward: 5′-GCCGCATCTTCTTTTGCGTCGC-3′, reverse: 5′-TCCCGTTCTCAGCCTTGACGGT-3′.14 The level of miR-302a-3p was evaluated using the comparative Ct method (ΔΔCt) and normalized by the GAPDH level.
Cell culture and transfection
The human HCC cell lines HepG2 and Huh7 were both obtained from the American Type Culture Collection (ATCC; Manassas, VA, USA) and cultured in DMEM supplemented with 10% fetal bovine serum (FBS; Thermo Fisher Scientific) and 1% penicillin/streptomycin at 37°C with 5% CO2.
The miR-302a-3p mimics and miR-negative control were ordered from GenePharma (Shanghai, People’s Republic of China). All the transfections were performed using Lipofectamine 2000 and then incubated in the incubator for 6 h. The cell culture medium was then replaced with the fresh medium containing 10% FBS. After cultured for 24–48 h, cells were subjected to functional assays.
Proliferation assay
MTT assay was used to measure the proliferation of HCC cells. Briefly, transfected cells were seeded onto a 96-well plate at a density of 1,000 cells/well in triplicate and cultured in DMEM containing 10% FBS. At the indicated time points (24, 48, 72, and 96 h), 20 μL of 5 mg/mL MTT was added to each well, and the cells were incubated for another 4 h at 37°C. Then, 200 μL of dimethyl sulfoxide (DMSO) was added per well to the cultured cells to dissolve the crystals. The absorbance was measured at 490 nm with a reference wavelength at 655 nm using a microplate reader (Bio-Rad Laboratories Inc., Hercules, CA, USA). The experiment was repeated three times independently. All the data were analyzed and plotted using GraphPad Prism 5.0 software (GraphPad Software, Inc., La Jolla, CA, USA).
Migration and invasion assays
Transwell chambers (8 μM, 24-well plate; EMD Millipore, Billerica, MA, USA) were used to measure the migration ability of HCC cells. Briefly, 2×104 cells were seeded into the upper chambers containing serum-free DMEM. The lower chamber was supplemented with 500 μL DMEM containing 10% FBS. After 24 h culture in the incubator, the membrane was carefully cleaned using a cotton swab to remove non-migrated cells, and cells that located on the bottom surface were fixed with 1% paraformaldehyde and then stained with 0.1% crystal violet. The invaded cell numbers were quantified by counting using a light microscope from five random fields for each well.
For the invasion assay, the transwell chambers were pre-coated with 50 μg Matrigel (BD Biosciences, San Jose, CA, USA), and then, 5×104 cells were seeded into each chamber. The subsequent procedures were same as those for migration analysis. All experiments were performed in triplicate and repeated for at least three times.
Plasmid, transfection, and luciferase activity assays
The PRKACB plasmid was purchased from Addgene, Cambridge, MA, USA and subcloned into pGL3 luciferase vector; the mutant of PRKACB was constructed by site-specific mutagenesis strategy.
HepG2 cells were seeded into a 24-well plate at the density of 1×104 cells/well. Cells were transfected with both miR302a-3p and PRKACB plasmids using Lipofectamine 2000 transfection reagent.15 Briefly, a blank vector was introduced as the control of miR302a-3p, which was also co-transfected with either PRKACB-wild type (WT) or PRKACB-mutant plasmids containing firefly luciferase. The value of relative luciferase activity was evaluated by a dual luciferase assay kit (Promega Corporation, Fitchburg, WI, USA) according to the manufacturer’s instructions.
Western blot
The Western blot was conducted to test the protein expression levels as described by others.16 Briefly, cultured cells were lysed using radioimmunoprecipitation assay (RIPA) buffer supplemented with protease inhibitor cocktails (Hoffman-La Roche Ltd., Basel, Switzerland). After determining the extracted protein concentration by a Pierce BCA protein quantification kit (Thermo Fisher Scientific), 20 μg of total proteins were separated by 12% sodium dodecyl sulfate polyacrylamide gel electrophoresis (SDS-PAGE) and transferred to nitrocellulose membranes. After blocking with 5% bovine serum albumin in Tris Buffered Saline with Tween 20, the nitrocellulose membranes were incubated with primary antibodies at 4°C overnight. Corresponding horseradish peroxidase (HRP)-conjugated secondary antibodies were added and allowed for another 1 h incubation at room temperature, and the immunoreactivity was finally detected by using X-ray film.
Statistics
Statistical analyses were carried out by using SPSS 20.0 (IBM Corporation, Armonk, NY, USA) and GraphPad Prism 5.0 software. Chi-square test was used to compare the statistical difference of two groups. OS time was defined as the period from the date of surgery to death or the end of follow-up (July 2017). The OS curves were plotted by the Kaplan–Meier method and compared by the logrank test. Univariate and multivariate Cox regression analyses were performed to explore the clinical significance of different variables. After confirming the distribution normality using the Shapiro–Wilk test and the variance homogeneity by the Levene’s test (all P>0.05), data from cellular experiments were analyzed using the Student’s t-test to assess the differences between groups. All data were represented as mean±SD, and P<0.05 was considered as statistically significant.
Results
Patients’ information
We enrolled 111 HCC patients in this study; all patients were pathologically diagnosed with HCC, and none of them received any antitumor therapy before surgery resection. All the surgery treatment was R0 resection. The median age of the patients at the time of surgery was 47 years. Most of the patients were male (91/111, 82.0%). As all the patients were collected in China, where hepatitis virus infection is the major cause of HCC, we thus retrieved the information about HBV infection. Accordingly, 87 (78.4%) patients were tested with HBV positive at the time of diagnosis. In all, 72 (64.9%) patients showed a higher serum AFP level (>400 U/mL). The information about the number of primary HCC and the largest tumor diameter was also collected. Most of the patients suffered from single tumor (72/111, 64.9%), and 62.2% patients had a tumor diameter >5.0 cm. Additionally, 31 (27.9%) patients showed tumor infiltration in portal vein, and 67 (60.4%) patients were classified to have TNM stage III/IV (Table 1).
  | Table 1 Clinical characteristics of the HCC patients |
miR-302a-3p is downregulated in HCC tissues and correlated with tumor progression
We tested the tissue expression levels of miR-302a-3p by RT-qPCR in both HCC tumor tissues and adjacent normal liver tissues. Most of the patients (74/111, 66.7%) showed a lower miR-302a-3p level in HCC tissues compared with that in normal liver tissues (Figure 1A and B). The chi-square test was next conducted to compare differential expression of miR-302a-3p in various subgroups. As a result, a lower miR-302a-3p level was correlated with multiple tumor involvement (P=0.003) and a larger tumor size (P<0.001). In addition, patients with advanced tumor stages were characterized with lower miR-302a-3p levels (P=0.028; Table 2), suggesting the possible tumor-suppressing role of miR-302a-3p in HCC development.
  | Table 2 Correlations between miR-302a-3p level and clinicopathological variables |
Low miR-302a-3p is an independent biomarker for predicting a poorer OS of HCC
To explore the role of miR-302a-3p in HCC prognosis, all the retrieved clinicopathological characteristics were subjected to Kaplan–Meier survival analyses (Figure 2 and Table 3). According to the univariate analysis and logrank test, prognostic factors included tumor number (P=0.006), tumor diameter (P=0.015), portal vein invasion (P=0.010), and TNM stage (P<0.001). Importantly, patients with higher miR-302a-3p levels also showed better clinical outcomes than those with lower miR-302a-3p levels (3-year OS 58.7% vs 28.2%; mean OS 38.8±3.2 vs 25.6±1.7 months; P=0.002). In contrast, there was no significant correlation between OS and patients’ age, sex, HBV infection, or serum AFP level.
  | Table 3 Kaplan–Meier survival analysis for HCC patients |
The variables with statistical significance found by univariate analysis were next subjected to a Cox regression model to explore their independent prognostic effect (Table 4). Both the TNM stage (HR=2.339, 95% CI=1.748–4.290, P=0.024) and miR-302a-3p level (HR=0.480, 95% CI=0.249–0.894, P=0.039) were identified as independent prognostic factors. Taken together, clinical data showed that miR-302a-3p is a tumor suppressor in HCC and can help predict patients’ OS.
  | Table 4 Multivariate analysis of HCC patients |
miR-302a-3p inhibits proliferation, migration, and invasion capacities of HCC cells
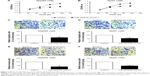
We also performed cellular experiments in two human HCC cell lines, HepG2 and Huh7. After overexpressing miR-302a-3p mimics, both HCC cell lines showed a decreased cell proliferation pattern (Figure 3A and B) as revealed by MTT assays. In addition, the cell migration ability was tested by transwell strategy, which showed that miR-302a-3p can inhibit HCC cell migration (Figure 3C and D). Finally, the Matrigel transwell assay was performed to investigate whether miR-302a-3p had an effect on cell invasion. According to our data, transfection of miR-302a-3p mimics remarkably inhibited the tumor cell invasion (Figure 3E and F), demonstrating its direct role in suppressing HCC progression.
miR-302a-3p targets PRKACB protein, thus inhibiting Src and CREB activation
We next aimed to explore the underlying mechanism of miR-302a-3p in inhibiting tumor progression of HCC. By using the TargetScan tool (www.targetscan.org), we found that PRKACB, the catalytic subunit beta of PKA kinase, possesses a potential binding site with miR-302a-3p (Figure 4A), which is consistent with a most recent study.17 Furthermore, we verified that miR-302a-3p can directly regulate the transcription of PRKACB by using luciferase assays (Figure 4B). In contrast, after mutating the binding sites on PRKACB, miR-302a-3p co-transfection showed little effect on its luciferase activity. Western blot results also showed that miR-302a-3p overexpression can inhibit the expression of PRKACB (Figure 4C). Additionally, the activation of PKA downstream effectors such as Src and CREB oncoproteins was also inhibited by miR-302a-3p. Considering the regulatory role of Src and CREB on cancer progression,18,19 we proposed a signaling network on the PRKACB-dependent effect of miR-302a-3p on inhibiting HCC proliferation and invasion (Figure 4D).
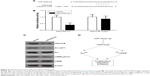  | Figure 4 miR-302a-3p directly suppresses PRKACB signaling. |
Discussion
Accumulating evidence highlighted the critical role of miRNAs in tumorigenesis and tumor development. miR-302 was first reported to be downregulated by 1,25(OH)2D3 treatment in several tumor cell lines, which resulted in a higher susceptibility to natural killer cell-mediated cytotoxicity.20 Although this datum revealed a possible oncogenic effect of miR-302a-3p, it did not illustrate its functional mechanisms from the molecular aspect.
Additionally, later studies reported that miR-302a suppressed the endothelial–mesenchymal transition of endothelial cells, subsequently playing tumor suppressing roles by indirectly inhibiting HCC progression.12 However, both the two studies mentioned earlier were based on “bench” findings, and the clinical significance of miR-302 was not recognized until 2015. Oksuz et al21 implied that serum miR-302 could be used as a biomarker for predicting occurrence of cirrhosis and HCC in HCV-positive patients. Moreover, miR-302a-3p showed a direct inhibitory effect on the proliferation and invasion of glioma cells,22 suggesting its role as a suppressor in solid tumors. Furthermore, it was recently reported that a low expression of miR-302a-3p was correlated with poorer survival of gastric cancer patients.11 Therefore, we sought to investigate the clinical significance and functional mechanisms of miR-302a-3p in HCC.
According to our data, miR-302a-3p was significantly downregulated in HCC tissues compared with adjacent liver tissues. Clinical data revealed its statistical correlation with tumor size and TNM stage. We also demonstrated its significance on predicting HCC survival by univariate and multivariate analyses. Furthermore, our results confirmed that miR-302a-3p can directly inhibit the proliferation, migration, and invasion processes of HCC cells. Besides the predictive role, miR-302a-3p was also reported to enhance the drug sensitivity in both breast cancer cells.23,24 Therefore, it is highly likely that miR-302a-3p may also increase the chemotherapy sensitivity of HCC cells, which needs further evidence. Additionally, our results provided evidence on the direct inhibiting role of miR-302a-3p on the PRKACB, thus attenuating the PKA activity. The phosphorylation levels of Src and CREB, two well-known oncoproteins regulated by PKA, were also inhibited by miR-302a-3p overexpression. It is thus reasonable that miR-302a-3p may suppress HCC progression at least partially by impairing PKA signaling.
To the best of our knowledge, this is the first study on tissue expression and clinical significance of miR-302a-3p in HCC. The inhibitory effect of miR-302a-3p on HCC progression deserves more attention, and it may be an invaluable direction for novel therapy development.
Conclusion
Our data showed that low miR-302a-3p level is an independent prognostic factor for predicting poorer OS of HCC patients, and miR-302a-3p suppressed HCC progression by directly inhibiting tumor cell proliferation and invasion capacities.
Acknowledgment
This study was supported by the Natural Science Foundation of China (No 81571718), Key Disciplines Group Construction Project of Pudong Health Bureau of Shanghai (No PWZxq2014-12), Science and Technology Development Fund (No 14DZ1940605), Science and Technology Development Fund of Shanghai Pudong New Area (No PKJ2017-Y14), and Talents Training Program of Seventh People’s Hospital of Shanghai University of TCM (No XX2017-04).
Disclosure
The authors report no conflicts of interest in this work.
References
Siegel RL, Miller KD, Jemal A. Cancer statistics, 2018. CA Cancer J Clin. 2018;68(1):7–30. | ||
Perz JF, Armstrong GL, Farrington LA, Hutin YJ, Bell BP. The contributions of hepatitis B virus and hepatitis C virus infections to cirrhosis and primary liver cancer worldwide. J Hepatol. 2006;45(4):529–538. | ||
Maluccio M, Covey A. Recent progress in understanding, diagnosing, and treating hepatocellular carcinoma. CA Cancer J Clin. 2012;62(6):394–399. | ||
Lee JG, Kang CM, Park JS, et al. The actual five-year survival rate of hepatocellular carcinoma patients after curative resection. Yonsei Med J. 2006;47(1):105–112. | ||
Lewis BP, Burge CB, Bartel DP. Conserved seed pairing, often flanked by adenosines, indicates that thousands of human genes are microRNA targets. Cell. 2005;120(1):15–20. | ||
Krek A, Grün D, Poy MN, et al. Combinatorial microRNA target predictions. Nat Genet. 2005;37(5):495–500. | ||
Calin GA, Croce CM. MicroRNA signatures in human cancers. Nat Rev Cancer. 2006;6(11):857–866. | ||
Volinia S, Calin GA, Liu C-G, et al. A microRNA expression signature of human solid tumors defines cancer gene targets. Proc Natl Acad Sci U S A. 2006;103(7):2257–2261. | ||
Zheng G, Wang Y, Zhu X. Evaluation of miR-302c-3p as prognostic biomarkers in glioma patients. Eur Rev Med Pharmacol Sci. 2016;20(8):1521–1525. | ||
Gu D-H, Mao J-H, Pan X-D, et al. microRNA-302c-3p inhibits renal cell carcinoma cell proliferation by targeting Grb2-associated binding 2 (Gab2). Oncotarget. 2017;8(16):26334. | ||
Ma G, Li Q, Dai W, Yang X, Sang A. Prognostic implications of miR-302a/b/c/d in human gastric cancer. Pathol Oncol Res. 2017;23(4):899–905. | ||
Zhu K, Pan Q, Jia L-Q, et al. MiR-302c inhibits tumor growth of hepatocellular carcinoma by suppressing the endothelial-mesenchymal transition of endothelial cells. Sci Rep. 2014;4:5524. | ||
Pan X, Li D, Huo J, Kong F, Yang H, Ma X. LINC01016 promotes the malignant phenotype of endometrial cancer cells by regulating the miR-302a-3p/miR-3130-3p/NFYA/SATB1 axis. Cell Death Dis. 2018;9(3):303. | ||
Liu H, Xu Y, Zhang Q, et al. Prognostic significance of TBL1XR1 in predicting liver metastasis for early stage colorectal cancer. Surg Oncol. 2017;26(1):13–20. | ||
Zhang Q, Yuan L, Liu D, et al. Hydrogen sulfide attenuates hypoxia-induced neurotoxicity through inhibiting microglial activation. Pharmacol Res. 2014;84:32–44. | ||
Tan W, Pan M, Liu H, Tian H, Ye Q, Liu H. Ergosterol peroxide inhibits ovarian cancer cell growth through multiple pathways. Onco Targets Ther. 2017;10:3467. | ||
Irwandi RA, Khonsuphap P, Limlawan P, Vacharaksa A. miR-302a-3p regulates RANKL expression in human mandibular osteoblast-like cells. J Cell Biochem. 2018;119(6):4372–4381. | ||
Frame MC. Src in cancer: deregulation and consequences for cell behaviour. Biochim Biophys Acta. 2002;1602(2):114–130. | ||
Conkright MD, Montminy M. CREB: the unindicted cancer co-conspirator. Trends Cell Biol. 2005;15(9):457–459. | ||
Min D, Lv X, Wang X, et al. Downregulation of miR-302c and miR-520c by 1, 25 (OH) 2D3 treatment enhances the susceptibility of tumour cells to natural killer cell-mediated cytotoxicity. Br J Cancer. 2013;109(3):723–730. | ||
Oksuz Z, Serin MS, Kaplan E, et al. Serum microRNAs; miR-30c-5p, miR-223-3p, miR-302c-3p and miR-17-5p could be used as novel non-invasive biomarkers for HCV-positive cirrhosis and hepatocellular carcinoma. Mol Biol Rep. 2015;42(3):713–720. | ||
Wang Y, Wei Y, Tong H, et al. MiR-302c-3p suppresses invasion and proliferation of glioma cells via down-regulating metadherin (MTDH) expression. Cancer Biol Ther. 2015;16(9):1308–1315. | ||
Wang Y, Zhao L, Xiao Q, et al. miR-302a/b/c/d cooperatively inhibit BCRP expression to increase drug sensitivity in breast cancer cells. Gynecol Oncol. 2016;141(3):592–601. | ||
Zhao L, Wang Y, Jiang L, et al. MiR-302a/b/c/d cooperatively sensitizes breast cancer cells to adriamycin via suppressing P-glycoprotein (P-gp) by targeting MAP/ERK kinase kinase 1 (MEKK1). J Exp Clin Cancer Res. 2016;35(1):25. |
 © 2018 The Author(s). This work is published and licensed by Dove Medical Press Limited. The full terms of this license are available at https://www.dovepress.com/terms.php and incorporate the Creative Commons Attribution - Non Commercial (unported, v3.0) License.
By accessing the work you hereby accept the Terms. Non-commercial uses of the work are permitted without any further permission from Dove Medical Press Limited, provided the work is properly attributed. For permission for commercial use of this work, please see paragraphs 4.2 and 5 of our Terms.
© 2018 The Author(s). This work is published and licensed by Dove Medical Press Limited. The full terms of this license are available at https://www.dovepress.com/terms.php and incorporate the Creative Commons Attribution - Non Commercial (unported, v3.0) License.
By accessing the work you hereby accept the Terms. Non-commercial uses of the work are permitted without any further permission from Dove Medical Press Limited, provided the work is properly attributed. For permission for commercial use of this work, please see paragraphs 4.2 and 5 of our Terms.



 refers to the phosphate group in biochemistry concept.
refers to the phosphate group in biochemistry concept.