Back to Journals » OncoTargets and Therapy » Volume 12
microRNA-26a-5p Promotes Proliferation and Migration of Osteosarcoma Cells by Targeting HOXA5 in vitro and in vivo
Authors Yu T, Chen D, Zhang L, Wan D
Received 23 September 2019
Accepted for publication 5 December 2019
Published 30 December 2019 Volume 2019:12 Pages 11555—11565
DOI https://doi.org/10.2147/OTT.S232100
Checked for plagiarism Yes
Review by Single anonymous peer review
Peer reviewer comments 3
Editor who approved publication: Dr Federico Perche
Tianhua Yu,1 Dexin Chen,2 Lei Zhang,3 Daqian Wan1,4,5
1Department of Orthopedics, Orthopedic Institute of Harbin, The Fifth Hospital in Harbin, Harbin, People’s Republic of China; 2School of Materials Engineering, Shanghai University of Engineering Science, Shanghai, People’s Republic of China; 3Department of Orthopedics, The First Affiliated Hospital of Shandong First Medical University, Shandong, People’s Republic of China; 4Key Laboratory of Spine and Spinal Cord Injury Repair and Regeneration, Ministry of Education of the People’s Republic of China, Shanghai, People’s Republic of China; 5Department of Orthopedics, Tongji Hospital Affiliated to Tongji University School of Medicine, Shanghai, People’s Republic of China
*These authors contributed equally to this work
Coresponding author: Daqian Wan; Lei Zhang Email [email protected]; [email protected]
Background: Osteosarcoma is the most common primary malignant tumor of bone. However, the underlying pathogenic mechanisms are still unclear. miR-26a was an endogenous non-coding small RNAs that have been showed to play a critical role in regulating varieties of biological and pathological processes. In this study, we will investigate the function of miR-26a-5p in osteosarcoma cells.
Methods: In this study, we explored the role of miR-26a-5p in osteosarcoma cell lines using qPCR, detected the proliferation, cell cycle and cell migration by CCK-8, PI and transwell.
Results: We found that compared with noncancerous cells, miR-26a-5p was highly expressed in osteosarcoma cell lines, especially in U2OS cells. Overexpression of miR-26a-5p promotes cell proliferation, cell cycle, and cell migration, but inhibits cell apoptosis. But down-regulation of miR-26a-5p in U2OS cells exhibits opposite effects. We also confirmed that miR-26a-5p directly targets HOXA5 in U2OS cells. Overexpression of HOXA5 reversed the effect of miR-26a-5p on cell proliferation, migration, and apoptosis. Besides, we showed in that knock-down of miR-26a-5p or overexpression of HOXA5 increased cell sensitivity to chemotherapeutic drug paclitaxel.
Conclusion: These findings indicate that highly expressed miR-26a-5p in osteosarcoma cells, and promotes proliferation and migration, but inhibits apoptosis of osteosarcoma cells by targeting HOXA5 which suggest that miR-26a-5p could serve as a novel therapeutic target for osteosarcoma.
Keywords: osteosarcoma MicroRNA, proliferation, migration, apoptosis
Introduction
Osteosarcoma is the most common primary malignant bone tumors affecting long bones in childhood and adolescence.1 Despite newly-developed multi-agent chemotherapy and gradually improved surgical techniques, the overall survival rate since the 1970s remains only approximately 60%.2 Till now, researches have found multiple and complex genomic aberrations in osteosarcoma, which is characterized by high number of structural variants with relatively small numbers of single nucleotide variants.3 But mechanisms underlying disease progression are still missing, and a better understanding of these mechanisms is essential to improve treatment options and patient outcomes.
microRNAs (miRNAs) are non-coding RNAs that play an essential role in regulating gene expression in a post-transcriptional manner.4–7 By base pairing to 3` untranslated region (3`UTR) of target mRNAs, miRNAs repress translation and/or lead to miRNA degradation. Abnormally expressed miRNAs were found to be involved in tumor development and progression, including osteosarcoma. Global microarray analyses were carried out and identified 177 miRNAs that were differentially expressed in human osteosarcoma cell lines relative to normal bone cells.8 Among which, miR-26a and miR-26b were further found to be up-regulated in tumor tissues and serum of osteosarcoma patients compared with healthy controls.9 These researches not only indicate that miR-26a and −26b could be served as diagnostic marker of osteosarcoma but also suggest that these two miRNAs might play important roles in osteosarcoma development and progression. Recently the role of miR-26a in osteosarcoma has been investigated.10–14 They found that overexpression of this miRNA accelerates cell cycle progression and inhibits apoptosis of MG63 and U2OS cells, two osteosarcoma cell line. However, the role of miR-26a-5p in osteosarcoma is completely unknown.
In this research, we investigated the possible role of miR-26a-5p in osteosarcoma and found that this miRNA promotes cell proliferation, migration, but inhibiting apoptosis. We further explored the underlying mechanism by detecting its target gene HOXA5.15,16 Our study may provide a novel mechanism and potential therapeutic target for osteosarcoma.
Materials and Methods
Cell Culture, Transfection, and Chemotherapeutic Agent Treatment
Human MSCs were donated by patients who provided written informed consent prior to the commencement of the study, and the ethical and legal approval was provided by the Institutional Ethics Committee of the Fifth hospital in Harbin before the study. The experiment was performed following guidelines and regulations of the Institutional Ethics Committee of the Fifth hospital in Harbin and other relevant national guidelines and regulations. Osteosarcoma cell lines (U2OS, Saos-2, and MG63), human MSCs, fibroblast cell line HFF-1, and osteoblast cell line hFOB1.19 were purchased from the Cell Bank of the Chinese Academy of Sciences (Shanghai, China). Cells other than hFOB1.19 were maintained in Dulbecco’s Modified Eagle’s Medium (DMEM, Hyclone, Logan City, Utah, USA) with 10% FBS (Gibco, Grand Island, New York, USA) and 1% penicillin and streptomycin (Hyclone, Logan City, Utah, USA) at 37°C culture conditions of 5% CO2. The hFOB1.19 cell line was maintained in DMEM with 15% FBS, 1% penicillin and streptomycin and 0.3 mg/mL G418 (Sigma, St. Louis, MO, USA) at 34.5°C culture conditions of 5% CO2. For the experiments, confluent cells were removed using 0.25% trypsin containing 10 mM EDTA (Hyclone, Logan City, Utah, USA), resuspended in antibiotic-free growth medium and plated onto six-well plates at a density of 2.0 × 105 cells per well (if not mentioned). For transfection assay, LipofectamineTM2000 (Invitrogen, USA) was used according to the Manufacturer’s Instruction. mimic-26a-5p or inhibitor-26a-5p (Ribobio Co., LTD, Guangzhou, China) were transfected at the concentration of 200 nM. For chemotherapeutic agent sensitivity assay, cells were transfected with miRNA mimic and inhibitor, and HOXA5 overexpression vector before being treated with 0.1μM Paclitaxel 12 hrs after treatment, apoptotic cells were detected by Annexin V assay.
RNA Isolation and Real-Time PCR
Total RNA from bone tissues or cells was extracted with TRIzol Reagent (Invitrogen, Mulgrave, Australia) according to the manufacturer’s instructions. First-strand cDNA was synthesized from 1 μg of total RNA by incubating for 1 hr at 42°C with Superscript III reverse transcriptase (Invitrogen, Mulgrave, Australia) following oligo (dT) priming. After reverse transcription reaction, qRT-PCR was performed by LightCycler480 system (Roche, Mannheim, Germany) using SYBR Premix Ex TaqTM (Takara, Dalian, China) according to the manufacturer’s instructions. miRNA amplifications were normalized by U6, mRNA amplifications were normalized by GAPDH. Data were analyzed using the comparison Ct (2−△△Ct) method and expressed as fold change compared to respective control. Each sample was analyzed in triplicate. The primer sequences used in this study were as follows: GAPDH: forward, 5ʹ-GAAAGCCTGCCGGTGACTAA-3ʹ; reverse, 5ʹ-AGGAAAAGCATCACCCGGAG-3ʹ; HOXA5: forward, 5ʹ-ATGCGCAAGCTGCACATAAG-3ʹ; reverse, 5ʹ- CGGGTCAGGTAACGGTTGAA-3ʹ.
Cell Proliferation, Cell Cycle, Migration and Apoptosis Assay
For proliferation assay, U2OS cells were seeded at a density of 4000 cells/well in 96-well plates, cell proliferation was monitored after indicated time points by MTT assay [3-(4, 5-dimethylthiazol-2-yl)-2, 5-diphenyltetrazolium bromide, a yellow tetrazole]. Cell cycle was detected using Cell Cycle and Apoptosis Analysis Kit (Beyotime Biotechnology, Shanghai, China) according to manufacture`s instructions. Briefly, cells were collected, washed in ice-cold PBS, and then fixed in 70% ethanol for 2 hrs at 4°C. After fixation, cells were washed in PBS, and then incubated in propidium iodide staining buffer for 30 mins at 37°C, following by flow cytometry detection. Cell migration was analyzed in 6.5-mm cell culture transwell inserts (8-μm pore size, Costar). Fifty-thousand cells were seeded in the upper chamber. After 24 hrs, the migrated cells that attached at the bottom of the lower chamber were counted. The data are expressed as the mean number of migrated cells per high power field.
For apoptosis assay, Dead Cell Apoptosis Kit with Annexin V Alexa Fluor™ 488 & Propidium Iodide (PI) kit (Life Technologies, Carlsbad, USA) was used according to manufacture`s instructions. Briefly, cells were collected in 100 μL binding buffer at density of 1×106 cell/mL, before adding 5 μL Alexa Fluor 488-annexin V and 1 μL PI. After incubating for 15 mins in the dark, 300 μL binding buffer was added to stop reaction. Cells then detected by flow cytometry.
Western Blot Analysis
For Western blot analysis, cells were lysed in lysis buffer (50 mM Tris-HCl, pH 7.4, 150 mM NaCl, 0.1% SDS, 1% Nonidet P-40, 1 mM PMSF) and protease inhibitor cocktail (10 mg/mL leupeptin, 10 mg/mL pepstatin A, and 10 mg/mL aprotinin) on ice for 30 mins. Protein fractions were collected by centrifugation at 15,000 g at 4°C for 10 mins and then subjected to 10% SDS-PAGE and transferred to polyvinylidene difluoride (PVDF) membranes. The membranes were blocked with 5% BSA and incubated with specific antibodies overnight at 4°C. A horseradish peroxidase–labeled secondary antibody was added and visualized using the enhanced chemiluminescence detection system (Millipore, Billerica, MA) as recommended by the manufacturer. Immunoreactive bands were quantitatively analyzed in triplicate by normalizing the band intensities to beta-actin on scanned films with Alpha Image software. Primary antibodies used in this study were human HOXA5 Rabbit mAb (1:1000) and beta-actin Rabbit mAb (1:1000, all purchased from Cell Signaling Technology, Inc).
HOXA5 3′ UTR Cloning and Luciferase Assay
HOXA5 mRNA 3ʹUTR containing the miR-26a-5p-binding sequences were amplified by PCR from human genomic DNA. Binding-region mutations were achieved using a QuikChange Site-Directed Mutagenesis Kit (Stratagene) following the manufacturer’s instructions. Luciferase constructs plasmids were co-transfected with pRL-TK Renilla luciferase plasmid (Promega, USA) into U2OS cells by Lipofectamine 2000 (Invitrogen). Luciferase assays were performed with the dual-luciferase reporter assay system (Promega) according to the manufacturer’s instructions. Luminescent signals were quantified by luminometer (Glomax, Promega), and each value from the Renilla luciferase construct was normalized by Firefly luciferase.
Lentiviral-Mediated HOXA5 Over-Expression
HOXA5 cDNA was cloned from U2OS total cDNA by following primers: forward: 5`-CCGCTCGAGATGAGCTCTTATTTTGTAAACT-3`, reverse: 5`- CGCGGATCCTCAGGGACGGAAGGCCCCT-3`. After purification, HOXA5 cDNA was subcloned into xhol and BamHlsite of pLVX-IRES-Puro plasmid. For virus packaging, 2 μg HOXA5 over-expression plasmid was co-transfected with 1.5 μg gpMD2 into 293FT cells. Forty-eight hours after transfection, supernatant was collected and filtrated for treatment of U2OS cells.
Statistical Analyses
All numerical data are expressed as the mean±S.D. Statistical differences among groups were analyzed by one-way analysis of variance with a post-hoc test (after normalization to baseline in the hindlimb-unloading study) to determine group differences in the study parameters. All statistical analyses were performed with SPSS software, version 13.0. Statistical differences between two groups were determined by the Student’s t test. P < 0.05 was considered statistically significant.
Results
miR-26a-5p Is Highly Expressed in Osteosarcoma Cell Lines
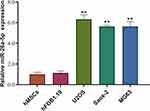
To investigate the possible roles that miR-26a-5p might play in osteosarcoma, we first detected its expression level in osteosarcoma cell lines U2OS, Saos-2 andMG63, a chondrosarcoma cell line. Human MSCs and osteoblast cell line hFOB1.19 were used as control. Our result shows that miR-26a-5p is highly expressed in every tested osteosarcoma cell lines compared to control cells, especially in U2OS (Figure 1). This result indicates that miR-26a-5p might be involved in the progression of osteosarcoma. Next, we focus on U2OS to further investigate the role of miR-26a-5p in osteosarcoma cells.
miR-26a-5p Promotes the Proliferation, Migration, but Inhibits Apoptosis of U2OS Cells
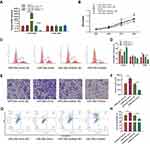
To investigate the molecular function of miR-26a-5p in U2OS, we transfected U2OS with miRNA mimic and inhibitor, respectively. Twenty-four hours after transfection, the mRNA levels of miR-26a-5p and miR-26a were detected by qRT-PCR, which shows that mimic and inhibitor significantly elevated and down-regulated the levels of miR-26a-5p but not miR-26b, respectively (Figure 2A). Next, we detected the effect of miRNA mimic and inhibitor on the cell proliferation, migration, and apoptosis of U2OS. MTT assay shows that miR-26a-5p mimic significantly promotes cell proliferation, while transfection of miR-26a-5p inhibitor exhibits opposite effect (Figure 2B). FCM assay shows that miR-26a-5p mimic increased the numbers of S and G2/M phase cells, while miR-26a-5p inhibitor decreased them (Figure 2C and D). These results indicate that miR-26a-5p promotes cell cycle and cell proliferation. Next, we performed transwell assay to detect the effect of miR-26a-5p on cell migration. U2OS cells that transfected with miR-26a-5p mimic showed greater migration ability. On the contrary, cells transfected with miR-26a-5p inhibitor showed lower migration rate than control cells (Figure 2E and F). To detect the effect of miR-26a-5p on cell apoptosis, U2OS cells were transfected miRNA mimic and inhibitor, respectively, before being detected by Annexin V assay. Forty-eight hours after transfection, we found that miR-26a-5p mimic does not significantly change cell apoptotic level, but miR-26a-5p inhibitor greatly promotes cell apoptosis (Figure 2G and H). Taken together, these results show that miR-26a-5p promotes cell proliferation, migration, but inhibit apoptosis of U2OS, indicating that highly expressed miR-26a-5p in osteosarcoma might positively correlate with carcinoma.
miR-26a-5p Targets HOXA5 in U2OS Cells
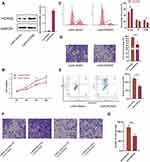
miRNAs function at the post-transcriptional level via degradation or translational inhibition of their target mRNAs by binding to the 3ʹ-untranslated region (3ʹUTR) of mRNA(4–7). To further illustrate molecular mechanisms of the effect of miR-26a-5p on U2OS, we performed bioinformatic analysis using TargetScan to screen for miR-26a-5p-targeting genes. Predicted genes including those involved in signal transduction, cell proliferation and differentiation, such as HOXA5 (Figure 3A). As we previously reported that HOXA5 might play a role in osteosarcoma, we choose to focus on this gene for further investigation. Targeting sequence of miR-26a-5p on HOXA5 mRNA is evolutionarily conserved across many vertebrate species (Figure 3A), indicating that HOXA5is a possible target of miR-26a-5p. To verify this, we first performed qPCR and Western blot assay to detect the expression level of HOXA5 in osteosarcoma cell lines (Saos-2, U2OS, and MG63), chondrosarcoma cell line (Sw1353) and control cells (HFF-1, hBMSC, and hFOB1.19). Consistent with the expression level of miR-26a-5p, both the mRNA and the protein level of HOXA5 were downregulated most significantly in U2OS cells (Figure 3B), suggesting miR-26a-5p might target HOXA5 in U2OS. To detect if miR-26a-5p can directly target HOXA5, we constructed luciferase reporters that contain wild type HOXA5 3`UTR sequence (HOXA5 UTR) or mutant sequence of miR-26a-5p binding sites (mut UTR), and then co-transfected with miR-26a-5p mimic and inhibitor in U2OS cells. The results of luciferase activity demonstrated that miR-26a-5p mimic greatly inhibited luciferase reporter activity, while miR-26b inhibitor elevated it but not mut UTR reporter (Figure 3C). Consistent with the luciferase reporter assays, qPCR and Western blot analysis also showed that miR-26b mimic or inhibitor does not significantly change HOXA5 mRNA level, but miR-26b mimic greatly decreased HOXA5 protein level in U2OS cells (Figure 3D). These results confirmed that HOXA5 was a direct target of miR-26b in U2OS cells.
HOXA5 Inhibits Proliferation and Migration, but Promotes Apoptosis of U2OS Cells
To demonstrate that miR-26a-5p function in U2OS through direct targeting HOXA5, we next detected the effect of HOXA5 on cell proliferation, migration, and apoptosis. As the expression level of HOXA5 in U2OS is too low, we over-expressed it by lentivirus-mediated transfection (Figure 4A). As expected, HOXA5 gain-of-function greatly inhibited proliferation (Figure 4B), cell cycle (Figure 4C), and cell migration (Figure 4D), as indicated by MTT, FCM and transwell assay, respectively. However, HOXA5 over-expression in U2OS cells greatly promotes cell apoptosis (Figure 4E). Overexpression of HOXA5 could rescue the effects induced by miR-26a mimic in migration of osteosarcoma cells (Figure 4F and G). Our results show that the effects of HOXA5 on U2OS are consistent with that of miR-26a-5p, indicating that highly expressed miR-26a-5p in U2OS promotes proliferation, migration and inhibits apoptosis by, at least in part, targeting HOXA5.
miR-26a-5p Overexpression and Knockdown Regulates the Tumorigenesis of Osteosarcoma in vivo
To further confirm the effect of miR-26a-5p on osteosarcoma progression, xenograft experiments were performed. We seeded the U2OS cells into nude mice. At indicative time points, we measured the osteosarcoma volumes and found that miR-26a-5p overexpression promoted the tumorigenesis of osteosarcoma and miR-26a-5p knockdown decreased the proliferation of osteosarcoma in vivo (Figure 5). Consistently, miR-26a-5p knockdown led to reduced tumor weights; however, miR-26a-5p overexpression increased the tumor weights (Figure 5).
miR-26a-5p and HOXA5 Regulate the Sensitivity of U2OS to Chemotherapeutics
As our above results showed that highly expressed miR-26a-5p in U2OS has anti-apoptosis effect, we hypothesize that inhibit miR-26a-5p expression could increase the sensitivity of U2OS cells to chemotherapeutic. To verify its possible role in regulating chemotherapeutic sensitivity, U2OS cells were transfected with miR-26a-5p inhibitor, before being treated by paclitaxel. Annexin V assay shows that knock-down of miR-26a-5p greatly increased paclitaxel-induced cell apoptosis (Figure 6A). Consistently, HOXA5 over-expression also promoted paclitaxel-induced cell apoptosis (Figure 6B). These results indicate that miR-26a-5p antagonizes paclitaxel-induced cell apoptosis by targeting HOXA5, and that miR-26a-5p could serve as a therapeutic target to improve sensitivity of osteosarcoma to paclitaxel.
Discussion
Osteosarcoma (OS) is the most common primary malignant bone tumor, occurring frequently in adolescents and possessing a high malignant severity. With the development of therapy treatments in OS, the 5-year survival rate was increased to approximately 60-70%.17,18 However, like to most malignant tumor, recurrence, metastasis and chemotherapeutic drug resistance resulted in a poor prognosis for OS patients. At present, the molecular pathogenesis and etiology of OS remain unclear.
MicroRNA molecules have a variety of roles in numerous biological and pathological processes, including cell proliferation, differentiation, and apoptosis. These effects are exerted through post-transcriptional regulation of gene expression via base pairing with target mRNA 3′-untranslated regions (3′-UTRs). It was reported that miRNAs modulate almost 60% of protein-coding genes in humans. Dysregulation of miRNAs is known to be involved in tumorigenesis and progression in various types of tumors. However, elucidation of the potential roles of miRNAs in osteosarcoma remains in the early stage of development. Large-scale expression screen that compare miRNA levels in tumors versus normal tissues have identified hundreds of novel miRNAs that are involved in osteosarcoma (8). These miRNAs might be implicated in osteosarcoma tumorigenesis, progression, invasion, and metastasis of osteosarcoma tumor cells. Recent studies revealed that these miRNAs function as either tumor suppressors such as miR-193a-3p/-5p,19 miR-506,20 miR-497,21 and miR-133a,22 or oncogenes such as miR-26a,13 miR-21,23 and miR-26a,18 depending on the role of their target genes. In this study, we focus on miR-26a-5p to explore its possible function in osteosarcoma.
miR-26a and miR-26b were found to be highly expressed in osteosarcoma tissues8 as well as in peripheral serum.9 In this study, we also found that compared with normal cells such as hMSCs, HFF-1, and hFOB1.19, the expression level of miR-26a-5p is much higher in osteosarcoma cell lines such as Saos-2 and U2OS. However, its possible roles in osteosarcoma are not known. A recent study report that miR-26a overexpression promotes cell proliferation and inhibits cell apoptosis.10 We hypothesized that highly expressed miR-26a-5p might play the same role in osteosarcoma cells. Therefore, we performed gain- and loss-of-function assay by transfecting miR-26a-5p mimic and inhibitor, respectively, into U2OS cells. As expected, miR-26a-5p mimic greatly improved cell proliferation, cell cycle, and migration. On the contrary, miR-26a-5p inhibitor down-regulated cell proliferation and migration, but significantly improved cell apoptosis. These findings confirmed our hypothesis that miR-26a-5p positively correlated with osteosarcoma growth and development.
Interestingly, the miR-26 miRNA gene family has been shown to function during embryogenesis and to be aberrantly expressed in various malignancies, including lung cancer,24 colorectal cancer,25 acute myeloid leukemia,26 and oral cancer.27 These reports together with our study demonstrated the significant role of miR-26a-5p in cancer development. Underlying mechanisms of the effect of miR-26a-5p in cancer development could be explained by its target genes. In this study, we predict HOXA5 as a target of miR-26a-5p by using public miRNA database TargetScan and confirmed that miR-26a-5p directly target HOXA5 in U2OS cells, as indicated in luciferase reporter assay. Consistent with the expression levels of miR-26a-5p, both the mRNA and protein levels of HOXA5 are much lower in osteosarcoma cell lines, especially in U2OS cells. Besides, we also found that overexpression of HOXA5 leads to repressed cell proliferation and migration, but improved cell apoptosis. These findings indicate that the effect of miR-26a-5p on U2OS cell is through, at least in part, targeting HOXA5.
Homeobox genes comprise a family of regulatory genes that contain a common homeobox domain and act as transcription factors that play fundamental roles in the morphogenesis of vertebrate embryonic cells, providing regional information along the main body axis.28 HOXA5 is also found as a tumor suppressor. The role of HOXA5 in osteosarcoma is completely unknown. Other studies report that HOXA5 inhibits cell proliferation, promote cell differentiation and apoptosis in many kind of cancers, such as breast cancer,29 acute myeloid leukemia,30 lung cancer,31 and colorectal cancer.16,32 Recent study demonstrated that colorectal cancer stem cells remain their stemness through Wnt signaling pathway-mediated HOXA5 expression inhibition. Forced expression of HOXA5 promotes cancer stem cell differentiation by inhibiting Wnt signaling. Thus, the reciprocal feedback between HOXA5 and Wnt signaling plays an important role in regulating cancer stem cell phenotype. Our study indicates that HOXA5 may also play an important role in osteosarcoma development. However, the underlying mechanisms need more investigation.
Therapeutic strategy of osteosarcoma includes surgical resection and chemotherapy. However, drug resistance leads to poor prognosis for osteosarcoma patients. In this study, we found that down-regulation of miR-26a-5p increased Paclitaxel-induced U2OS cell apoptosis, indicating that miR-26a-5p could be as a target to increase osteosarcoma chemosensitivity. Gondi et al have found that uPA maintains the stemness of pancreatic cancer cells by inhibiting the expression of HOXA5, thereby promoting the gemcitabine resistance of pancreatic cancer cells.33 In our study, we found that miR-26a-5p also increased the resistance of osteosarcoma cells to paclitaxel by down-regulating the expression of HOXA5. Therefore, HOXA5 may be a new target for the treatment of tumor resistance. We also reported for the first time the role of miR-26a-5p in tumor resistance and its mechanism of action.
In conclusion, our study demonstrated that miR-26a-5p is highly expressed in osteosarcoma cells, which promotes cell proliferation, cell cycle, and cell migration, but inhibit cell apoptosis by targeting tumor repressor HOXA5. These findings indicate that miR-26a-5p could be as a potential therapeutic target of osteosarcoma.
Acknowledgments
This work was supported by grants from the Shandong Key Research and Development Plan (2019GSF107057), Shandong medical and health science and technology development program (No. 2017WS446).
Disclosure
The authors report no conflicts of interest in this work.
References
1. Heare T, Hensley MA, Dell’Orfano S. Bone tumors: osteosarcoma and Ewing’s sarcoma. Curr Opin Pediatr. 2009;21:365–372. doi:10.1097/MOP.0b013e32832b1111
2. Ferguson WS, Goorin AM. Current treatment of osteosarcoma. Cancer Invest. 2001;19:292–315. doi:10.1081/CNV-100102557
3. Chen X, Bahrami A, Pappo A, et al. Recurrent somatic structural variations contribute to tumorigenesis in pediatric osteosarcoma. Cell Rep. 2014;7:104–112. doi:10.1016/j.celrep.2014.03.003
4. Ambros V. The functions of animal microRNAs. Nature. 2004;431:350–355. doi:10.1038/nature02871
5. Kosik KS. MicroRNAs and cellular phenotypy. Cell. 2010;143:21–26. doi:10.1016/j.cell.2010.09.008
6. Bartel DP. MicroRNAs: genomics, biogenesis, mechanism, and function. Cell. 2004;116:281–297. doi:10.1016/S0092-8674(04)00045-5
7. Bartel DP. MicroRNAs: target recognition and regulatory functions. Cell. 2009;136:215–233. doi:10.1016/j.cell.2009.01.002
8. Namlos HM, Meza-Zepeda LA, Baroy T, et al. Modulation of the osteosarcoma expression phenotype by microRNAs. PLoS One. 2012;7:e48086. doi:10.1371/journal.pone.0048086
9. Zhang C, Yao C, Li H, Wang G, He X. Combined elevation of microRNA-196a and microRNA-196b in sera predicts unfavorable prognosis in patients with osteosarcomas. Int J Mol Sci. 2014;15:6544–6555. doi:10.3390/ijms15046544
10. Shang Y, Wang LQ, Guo QY, Shi TL. MicroRNA-196a overexpression promotes cell proliferation and inhibits cell apoptosis through PTEN/Akt/FOXO1 pathway. Int J Clin Exp Pathol. 2015;8:2461–2472.
11. Liu J, Mi B, Wang Y, et al. miR-26a suppresses osteosarcoma migration and invasion by directly targeting HMGA1. Oncol Lett. 2018;15:8303–8310. doi:10.3892/ol.2018.8359
12. Lu J, Song G, Tang Q, et al. MiR-26a inhibits stem cell-like phenotype and tumor growth of osteosarcoma by targeting Jagged1. Oncogene. 2017;36:231–241. doi:10.1038/onc.2016.194
13. Qu F, Li CB, Yuan BT, et al. MicroRNA-26a induces osteosarcoma cell growth and metastasis via the Wnt/beta-catenin pathway. Oncol Lett. 2016;11:1592–1596. doi:10.3892/ol.2015.4073
14. Tan X, Fan S, Wu W, Zhang Y. MicroRNA-26a inhibits osteosarcoma cell proliferation by targeting IGF-1. Bone Res. 2015;3:15033. doi:10.1038/boneres.2015.33
15. Teo WW, Merino VF, Cho S, et al. HOXA5 determines cell fate transition and impedes tumor initiation and progression in breast cancer through regulation of E-cadherin and CD24. Oncogene. 2016;35:5539–5551. doi:10.1038/onc.2016.95
16. Ordonez-Moran P, Dafflon C, Imajo M, Nishida E, Huelsken J. HOXA5 counteracts stem cell traits by inhibiting Wnt signaling in colorectal cancer. Cancer Cell. 2015;28:815–829. doi:10.1016/j.ccell.2015.11.001
17. Mirabello L, Troisi RJ, Savage SA. Osteosarcoma incidence and survival rates from 1973 to 2004: data from the surveillance, epidemiology, and end results program. Cancer. 2009;115:1531–1543. doi:10.1002/cncr.v115:7
18. Cho Y, Jung GH, Chung SH, Kim JY, Choi Y, Kim JD. Long-term survivals of stage IIb osteosarcoma: a 20-year experience in a single institution. Clin Orthop Surg. 2011;3:48–54. doi:10.4055/cios.2011.3.1.48
19. Pu Y, Zhao F, Cai W, Meng X, Li Y, Cai S. MiR-193a-3p and miR-193a-5p suppress the metastasis of human osteosarcoma cells by down-regulating Rab27B and SRR, respectively. Clin Exp Metastasis. 2016;33:359–372. doi:10.1007/s10585-016-9783-0
20. Yu Z, Zhang Y, Gao N, Wang X. Overexpression of miR-506 inhibits growth of osteosarcoma through Snail2. Am J Transl Res. 2015;7:2716–2723.
21. Ruan WD, Wang P, Feng S, Xue Y, Zhang B. MicroRNA-497 inhibits cell proliferation, migration, and invasion by targeting AMOT in human osteosarcoma cells. Onco Targets Ther. 2016;9:303–313. doi:10.2147/OTT
22. Chen G, Fang T, Huang Z, et al. MicroRNA-133a inhibits osteosarcoma cells proliferation and invasion via targeting IGF-1R. Cell Physiol Biochem. 2016;38:598–608. doi:10.1159/000438653
23. Lv C, Hao Y, Tu G. MicroRNA-21 promotes proliferation, invasion and suppresses apoptosis in human osteosarcoma line MG63 through PTEN/Akt pathway. Tumour Biol. 2016;37:9333–9342. doi:10.1007/s13277-016-4807-6
24. Yu SL, Lee DC, Sohn HA, et al. Homeobox A9 directly targeted by miR-196b regulates aggressiveness through nuclear Factor-kappa B activity in non-small cell lung cancer cells. Mol Carcinog. 2016;55:1915–1926. doi:10.1002/mc.22439
25. Guan X, Yi Y, Huang Y, et al. Revealing potential molecular targets bridging colitis and colorectal cancer based on multidimensional integration strategy. Oncotarget. 2015;6:37600–37612. doi:10.18632/oncotarget.v6i35
26. Das RP, Konkimalla VB, Rath SN, Hansa J, Jagdeb M. Elucidation of the molecular interaction between miRNAs and the HOXA9 gene, involved in acute myeloid leukemia, by the assistance of argonaute protein through a computational approach. Genomics Inform. 2015;13:45–52. doi:10.5808/GI.2015.13.2.45
27. Lu YC, Chang JT, Huang YC, et al. Combined determination of circulating miR-196a and miR-196b levels produces high sensitivity and specificity for early detection of oral cancer. Clin Biochem. 2015;48:115–121. doi:10.1016/j.clinbiochem.2014.11.020
28. Ford HL. Homeobox genes: a link between development, cell cycle, and cancer? Cell Biol Int. 1998;22:397–400. doi:10.1006/cbir.1998.0329
29. Chen H, Chung S, Sukumar S. HOXA5-induced apoptosis in breast cancer cells is mediated by caspases 2 and 8. Mol Cell Biol. 2004;24:924–935. doi:10.1128/MCB.24.2.924-935.2004
30. Strathdee G, Sim A, Soutar R, Holyoake TL, Brown R. HOXA5 is targeted by cell-type-specific CpG island methylation in normal cells and during the development of acute myeloid leukaemia. Carcinogenesis. 2007;28:299–309. doi:10.1093/carcin/bgl133
31. Wang XC, Tian LL, Wu HL, et al. Expression of miRNA-130a in nonsmall cell lung cancer. Am J Med Sci. 2010;340:385–388. doi:10.1097/MAJ.0b013e3181e892a0
32. Tan SH, Barker N. Stemming colorectal cancer growth and metastasis: HOXA5 forces cancer stem cells to differentiate. Cancer Cell. 2015;28:683–685. doi:10.1016/j.ccell.2015.11.004
33. Asuthkar S, Stepanova V, Lebedeva T, et al. Multifunctional roles of urokinase plasminogen activator (uPA) in cancer stemness and chemoresistance of pancreatic cancer. Mol Biol Cell. 2013;24:2620–2632. doi:10.1091/mbc.e12-04-0306
 © 2019 The Author(s). This work is published and licensed by Dove Medical Press Limited. The full terms of this license are available at https://www.dovepress.com/terms.php and incorporate the Creative Commons Attribution - Non Commercial (unported, v3.0) License.
By accessing the work you hereby accept the Terms. Non-commercial uses of the work are permitted without any further permission from Dove Medical Press Limited, provided the work is properly attributed. For permission for commercial use of this work, please see paragraphs 4.2 and 5 of our Terms.
© 2019 The Author(s). This work is published and licensed by Dove Medical Press Limited. The full terms of this license are available at https://www.dovepress.com/terms.php and incorporate the Creative Commons Attribution - Non Commercial (unported, v3.0) License.
By accessing the work you hereby accept the Terms. Non-commercial uses of the work are permitted without any further permission from Dove Medical Press Limited, provided the work is properly attributed. For permission for commercial use of this work, please see paragraphs 4.2 and 5 of our Terms.