Back to Journals » International Journal of General Medicine » Volume 14
microRNA-19a-3p and microRNA-376c-3p Promote Hepatocellular Carcinoma Progression Through SOX6-Mediated Wnt/β-Catenin Signaling Pathway
Authors Cao X, Zhang J, Apaer S, Yao G, Li T
Received 24 August 2020
Accepted for publication 2 December 2020
Published 13 January 2021 Volume 2021:14 Pages 89—102
DOI https://doi.org/10.2147/IJGM.S278538
Checked for plagiarism Yes
Review by Single anonymous peer review
Peer reviewer comments 2
Editor who approved publication: Dr Scott Fraser
Xinling Cao,1 Jingjing Zhang,2 Shadike Apaer,1 Gang Yao,1 Tao Li1
1Department of Liver Transplantation & Laparoscopic Surgery, The First Affiliated Hospital of Xinjiang Medical University, Urumqi, Xinjiang 830054, People’s Republic of China; 2Department of Nephrology, The First Affiliated Hospital of Xinjiang Medical University, Urumqi, Xinjiang 830054, People’s Republic of China
Correspondence: Tao Li
Department of Liver Transplantation & Laparoscopic Surgery, The First Affiliated Hospital of Xinjiang Medical University, No. 137 Liyushan South Road, Xinshi District, Urumqi, Xinjiang 830054, People’s Republic of China
Tel +86-18799190276
Fax +86-0991-4364529
Email [email protected]
Background: Recent researches have suggested that microRNA (miR)-19a-3p and miR-376c-3p might function as initiators in diverse cancers. Based on which, in this current study, we aimed to probe into the combined effects and mechanisms of miR-19a-3p and miR-376c-3p in hepatocellular carcinoma (HCC) cells.
Methods: Tumor tissues and adjacent normal tissues from 21 cases of HCC patients, HCC cell lines, and human normal liver cell lines were used in this study. RT-qPCR and Western blot were implemented to detect the expression of miR-19a-3p, miR-376c-3p, SOX6, and Wnt/β-catenin pathway-associated factors in HCC tissues and cells. The direct relationships between miR-19a-3p or miR-376c-3p and SOX6 were confirmed by luciferase activity assay. HCC cells were treated with miR-19a-3p inhibitor, miR-376c-3p inhibitor, or oe-SOX-6 to figure out their functions in HCC malignancy. The in vivo assays were conducted for the confirmation of in vitro results.
Results: In both HCC tissues and cells, miR-19a-3p and miR-376c-3p were highly expressed, and SOX6 was poorly expressed. Depleted miR-19a-3p or miR-376c-3p was found to result in retarded HCC development. Bioinformatics analysis and luciferase activity assay revealed that SOX6 was the common target gene of miR-19a-3p and miR-376c-3p. Overexpressed SOX6 was demonstrated to block the Wnt/β-catenin pathway, thereby slowing down HCC progression. The in vivo assays showed that suppressed miR-19a-3p or miR-376c-3p and elevated SOX6 could reduce the tumor volume and weight of nude mice.
Conclusion: This study suggests that miR-19a-3p/miR-376c-3p activates the Wnt/β-catenin pathway via targeting SOX6, contributing to promoted biological functions of HCC cells.
Keywords: hepatocellular carcinoma, microRNA-19a-3p, microRNA-376c-3p, SOX6, Wnt/β-catenin signaling pathway, malignant progression
Introduction
Hepatocellular carcinoma (HCC) is one of the most frequent types of cancers around the world and becomes a leading reason of cancer-related mortality due to its increasing incidence.1 Obesity, diabetes, excess alcohol intake, metabolic diseases, and infection with hepatitis B or C are the primary risk factors of HCC, and these factors contribute to progressive fibrosis and cirrhosis.2 The diagnostic and therapeutic methods have made great advancements, but the long-term prognosis of advanced HCC patients remains dismal because of the high metastasis and recurrence post-operation.3 In addition, HCC patients often display different clinical outcomes even with the same clinicopathologic features, suggesting the complicated cellular and molecular events involved in HCC development.4 Thus, it is extremely urgent to seek for the optimal biomarkers for early diagnosis and efficient therapy of HCC through clarifying the molecular mechanisms of HCC progression.
microRNAs (miRNAs) can complementarily bind to the 3ʹ untranslated region (UTR) of their target messenger RNAs (mRNAs), resulting in mRNA degradation or prevention of translation.5 It is reported that miRNAs could potentially function as new biomarkers for the diagnosis and prognosis of HCC.6 MiR-19a, which belongs to the miR-17-92 cluster, is located on chromosome 13q31.3.7 Some researchers have suggested that miR-19a is involved in HCC occurrence and development.4,8,9 miR-376c-3p has been found to be elevated in HCC tissues and predicts poor prognosis of HCC patients.10 Moreover, overexpression of miR-18a and miR-25, which belong to miR-17-92 and miR-25-106b clusters, respectively, was associated with poor survival of HCC patients and promoted proliferation in HCC cells.11 Yet, the combined functions of miR-19a-3p and miR-376c-3p, as well as their downstream target are scarcely explored in HCC progression. In this current study, dual-luciferase reporter gene assay confirmed the binding relationship between miR-19a-3p and sex determining region Y-related high mobility group box 6 (SOX6), or miR-376c-3p and SOX6, respectively. As a member of the D subfamily of the SOX transcription factors, SOX6 harbors a long 3ʹ-UTR that shares various miRNA targets and is of great significance for cell proliferation, metastasis, as well as apoptosis.12 For example, Guo et al have stated that SOX6 is a novel prognostic marker for HCC patients owing to the close association between poor expression of SOX6 and its poor prognosis.13 Considering this, our study elaborated a new miR-19a-3p/miR-376c-3p-SOX6 regulatory pathway in the development of HCC, aiming to provide potential biomarkers and therapeutic targets for HCC.
Materials and Methods
Ethics Statement
This research was ratified by the Medical Ethics Committee of The First Affiliated Hospital of Xinjiang Medical University. All patients signed the informed consent. The experimental procedures were implemented following the Guidelines for Care and Use of Laboratory Animal with the ratification of Ethics Committee of The First Affiliated Hospital of Xinjiang Medical University.
Clinical Sample
The liver biopsy tissues and adjacent normal tissues of 21 HCC patients (aged 46–73 years) were obtained from The First Affiliated Hospital of Xinjiang Medical University from June 2016 to June 2017. All HCC patients were diagnosed via pathological examination and had not received any radiotherapy and chemotherapy before enrollment. Additionally, all patients had no history of other cancers. The clinicopathologic characteristics of patients with HCC are presented in Table 1. The expression of miR-19a-3p, miR-376c-3p, and SOX6 in HCC patients was obtained from The Cancer Genome Atlas (TCGA) database (https://cancergenome.nih.gov/), and the patients were divided into two groups using the survival program (version 3.6.3, R) with the median gene expression as the criterion. Prognostic analysis was performed by Kaplan-Meier analysis.
 |
Table 1 Clinicopathologic Characteristics of Patients with HCC |
Microarray-Based Analysis
For the analysis of differentially expressed miRNAs and mRNAs, the corresponding probes (Agilent Technologies, Palo Alto, CA, USA) were recovered in a 50 mM SpotArray 24 Microarray Printing System (SSC). Total RNA was extracted with RNeasy Mini Kit (Qiagen, Valencia, CA, USA), and 30 μg of which was purified on 15% denaturing polyacrylamide gel. The reverse transcription kit (RR047A, Takara, Shiga, Japan) was used to generate complementary DNA (cDNA). Next, the cDNA was amplified by polymerase chain reaction (PCR) and labeled with miRNA Complete Labeling and Hyb Kit (Agilent). PCR product was supplemented with hybridization buffer (Ambion, Austin, TX, USA), and the solution was hybridized with the probe at 42°C overnight. ScanArray Express Microarray Scanner (PerkinElmer, Waltham, MA, USA) was utilized for scanning, and the obtained data were subjected to QC analysis and normalization. Differentially expressed genes were screened based upon the criteria of p < 0.01 and |log2 Fold change| > 1, and the expression heat map was plotted.
Cell Culture and Transfection
Human normal liver cell line THLE3 and HCC cell lines (Hep3B, American Type Culture Collection, Rockville, Maryland, USA; MHCC-97L and Huh7, Shanghai Zhongqiao Xinzhou Biotechnology Co., Ltd., Shanghai, China) were, respectively, seeded in Roswell Park Memorial Institute (RPMI)-1640 cell culture medium (Gibco, Grand Island, NY, USA) containing 10% fetal bovine serum (FBS), 100 U/mL penicillin and 100 μg/mL streptomycin. Cells were cultured at 37°C with 95% saturated humidity.
The cells were seeded onto a 6-well plate 24 h before transfection, and the transfection process was performed in the light of Lipofectamine 2000 (11668-019, Invitrogen, New York, CA, USA) upon reaching 30–50% confluence. The cells were treated with miR-19a-3p inhibitor, miR-376c-3p inhibitor, miR-19a-3p inhibitor + miR-376c-3p inhibitor, overexpressed (oe)-SOX6, oe-SOX6 + miR-19a-3p mimic, or oe-SOX6 + miR-376c-3p mimic, as well as their corresponding negative controls (NCs). The plasmids were available from Shanghai GenePharma Co., Ltd. (Shanghai, China). The cells were cultured in an incubator at 37°C, and the complete medium was renewed 6 h later. The subsequent experiments were carried out in cells after a 48-h culture.
Immunohistochemical Staining
Paraffin-embedded sections were dewaxed, dehydrated with gradient alcohol, treated with 3% methanol H2O2 for 20 min, and rinsed with distilled water for 2 min and 0.1 M phosphate-buffered saline (PBS) for 3 min. After retrieving with water bath in antigen repair solution, the tissues were reacted with normal goat serum blocking solution (C-0005, Shanghai Haoran Bio Technologies Co., Ltd., Shanghai, China). Afterwards, the tissues were cultured at 4°C overnight with the primary rabbit anti-human antibody against SOX6 (ab64946, 1:200, Abcam, Cambridge, UK) and secondary goat anti-rabbit antibody against IgG (ab6785, 1:1000, Abcam) at 37°C for 20 min. Subsequently, the tissues were reacted at 37°C for 20 min with streptavidin and biotinylated horseradish peroxidase complex (0343–10000U, Beijing Imunbio Inc., Beijing, China). Next, the tissues were developed by diaminobenzidine (ST033, Guangzhou Whiga Technology Co., Ltd., Guangzhou, Guangdong, China), counterstained for 1 min with hematoxylin (PT001, Shanghai Bogoo Biotechnology. Co., Ltd., Shanghai, China), treated with 1% ammonia, dehydrated with gradient alcohol, cleared with xylene, and blocked with neutral resin. The tissues were captured under a microscope (Carl Zeiss, Jena, Germany), and SOX6 was mainly expressed in the cytoplasm with yellowish brown staining observed.
Cell Counting Kit-8 (CCK-8) Assay
The cells with 48-h transfection were detached and resuspended, and the cell concentration was modulated to 1 × 105 cells/mL. The cells were seeded onto 96-well plates with 100 μL/well, and then routinely cultured overnight. After that, the cells were treated based upon the requirements of the CCK-8 kit (Beyotime, Shanghai, China). CCK-8 detection solution (10 μL) was appended at 0, 24th, 48th and 72nd h and incubated for 4 h. The optical density (OD) value at 450 nm was measured by a microplate reader (elx800, BioTek, Winooski, VT, USA), and the growth curve was plotted accordingly.
Transwell Assay
The mixture of Matrigel (356234, BD Biosciences, Franklin Lakes, NJ, USA) and serum-free medium (1:3) were added into the Transwell chamber at 50 μL/well. Matrigel was polymerized into gel in a 37°C incubator. Treated in serum-free medium for 12 h, the detached cells were resuspended to 1 × 105 cells/mL in serum-free medium. The culture medium with 10% FBS was appended in the basolateral chamber, and the cell suspension (100 μL) was supplemented into the apical chamber. The cells were placed in a cell incubator with 5% CO2 saturated humidity at 37°C for 24 h. The cells that did not invade the Matrigel membrane were gently wiped off with cotton swabs, fixed with 4% paraformaldehyde, and stained with crystal violet (Sigma-Aldrich, St. Louis, MO, USA). Migration assays were performed without the addition of Matrigel and the remaining steps were the same as for the invasion assays. The stained invasive and migratory cells were counted in 5 visual fields under an inverted light microscope (Carl Zeiss).
Flow Cytometry
The cell apoptosis was performed with Annexin V-fluoresce in isothiocyanate/propidium iodide (FITC/PI) apoptosis detection kit (MA0220, Dalian Meilun Biotech Co., Ltd., Dalian, China). In brief, the cells (2~5 × 105 cell/mL) were harvested and centrifuged at 500 g for 5 min, followed by reaction with 195 μL binding buffer and 5 μL annexin V-FITC in the dark for 10 min at room temperature. Afterwards, the cells were reacted with 10 μL PI (20 μg/mL) in the dark for 5 min at room temperature. Apoptosis was detected by a flow cytometer (attune NXT, Thermo Fisher Scientific, Waltham, MA, USA). Apoptosis rate = (number of apoptotic cells/total number of measured cells) × 100%.
Reverse Transcription Quantitative PCR (RT-qPCR)
Total RNA was extracted by RNeasy Mini Kit (Qiagen), and the cDNA was obtained based on a reverse transcription kit (RR047A, Takara). For miRNA detection, the miRNA First Strand cDNA Synthesis (Tailing Reaction) kit (B532451-0020, Sangon Biotech, Shanghai, China) was used for reverse transcription to obtain the cDNA. SYBR® Premix Ex TaqTM II (Perfect Real Time) kit (DRR081, Takara) was utilized for sampling, and the samples were subjected to RT-qPCR reaction in real-time fluorescent qPCR (ABI 7500, ABI, Foster City, CA, USA). The universal reverse primers for miRNA and forward primers for U6 (internal reference) were provided by miRNA first strand cDNA synthesis (Tailing Reaction) kit, and other primers were synthesized by Sangon (Table 2). U6 and glyceraldehyde phosphate dehydrogenase (GAPDH) were the internal reference of genes. The expression of products was tested by 2-ΔΔCt method.
 |
Table 2 Primer Sequence |
Dual-Luciferase Reporter Gene Assay
Binding sites of miR-19a-3p and miR-376c-3p to the SOX6-wild type (WT) and the SOX6-mutant type (MUT) of the SOX6 3ʹUTR were artificially synthesized. The pmiR-RB-REPORTTM plasmid was digested by restriction endonuclease. The synthetic target gene fragments WT and MUT were inserted into the pmiR-RB-REPORTTM vector Guangzhou RiboBio Co., Ltd. (Guangzhou, Guangdong, China). The empty plasmid was transfected as a control group. The luciferase reporter plasmids WT and MUT with correct sequence were used for subsequent transfection. The vectors containing MUT and WT were co-transfected into HEK293T cells (ATCC) with miR-19a-3p mimic, miR-376c-3p mimic or mimic NC, respectively. A renilla luciferase detection kit (YDJ2714, Shanghai Yuduo Biotechnology Co., Ltd., Shanghai, China) was utilized to determine the relative light unit (RLU). A dual-luciferase reporter analysis system (Promega Co, Madison, WI, USA) was used for analysis. Relative luciferase activity was calculated by the RLU value of renilla luciferase/the RLU value of firefly luciferase.
Western Blot Analysis
The cells were lysed with 1 mL cell lysis buffer (BC-WB-018, Nanjing BioChannel Biotechnology Co., Ltd., Nanjing, China) at 4°C for 0.5 h, and centrifuged for 20 min at 12,000 r/min (4°C) to remove the lipid layer. With the supernatant as the protein extract, the protein concentration of protein was detected by a bicinchoninic acid kit (20201ES76, Yeasen Biotech Co., Ltd., Shanghai, China). Next, the protein was separated by sodium dodecyl sulfate polyacrylamide gel electrophoresis and then electroblotted onto polyvinylidene fluoride membranes. Blocking with 5% skim milk powder at 4°C overnight, the membranes were incubated overnight at 4°C with specific antibodies, such as rabbit-anti SOX6 (1:1000, ab64946), wnt-5a (1:1000, ab229200), β-catenin (1:2000, ab6302), or GAPDH (1:1000, ab9485) (all from Abcam), as well as HRP-labeled goat anti-rabbit IgG (ab6721, 1:5000, Abcam) diluent at room temperature for 60 min, followed by washing and development. The gray value of the protein band was processed by Quantity One software. GAPDH was taken as the loading control.
In vivo Experiment
Lentivirus was utilized to infect Hep3B cells with inhibition of miR-19a-3p and miR-376c-3p and stable high expression of SOX6. A total of 25 BALB/c nude mice aged 4–6 weeks were randomly divided into five groups (n = 5 mice). Hep3B cells in exponential growth phase were collected resuspended. The cell suspension (100 μL, 1 × 107 cells/mL) was subcutaneously injected into the axillary subcutaneous tissues of nude mice. The tumor growth of nude mice was observed every day after injection. The short diameter (a) and long diameter (b) of the tumor were recorded weekly, and the tumor volume was calculated based on the formula a × b2/2. Thirty-five days later, the nude mice were euthanized by overdose sodium pentobarbital at 150 mg/kg. The tumors were dissected, and the weight of tumors was weighed. The tumor tissues of mice fixed with 4% paraformaldehyde, followed by paraffin-embedding, sectioning, and immunohistochemical staining analysis.
Statistics
All data were processed by SPSS 21.0 statistical software (IBM Corp., Armonk, NY, USA). The measurement data were depicted as mean ± standard deviation. After verifying that the data are normally distributed, parametric tests were conducted. The data between the two groups were compared by unpaired t-test and/or the data among multiple groups were compared by one-way analysis of variance (ANOVA) or two-way ANOVA. Significant difference was set at p < 0.05.
Results
miR-19a-3p and miR-376c-3p are Upregulated in HCC Tissues and Cells
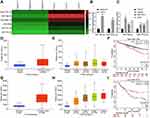
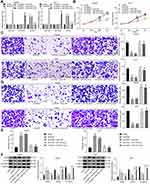
For detecting the differentially expressed miRNAs in HCC tissues and exploring the mechanism of HCC development, we performed the microarray-based analysis on HCC tissues and adjacent normal tissues of three patients (Figure 1A). The results suggested that miR-19a-3p and miR-376c-3p expression in HCC tissues was higher than that in the adjacent normal tissues. The high expression of miR-19a-3p and miR-376c-3p in HCC tissues was further verified by RT-qPCR detection (Figure 1B). In addition, RT-qPCR was also implemented for the detection of miR-19a-3p and miR-376c-3p expression in human normal liver cell line THLE3 and HCC cell lines (Hep3B, MHCC-97L, and Huh7). Similarly, miR-19a-3p and miR-376c-3p expression in HCC cell lines was elevated versus THLE3 cell line (p < 0.05; Figure 1C). These results imply that miR-19a-3p and miR-376c-3p are highly expressed in HCC tissues and cells. The expression of miR-19a-3p and miR-376c-3p was detected in TCGA database, and miR-19a-3p was found to be highly expressed in HCC patients (p < 0.05; Figure 1D). Moreover, the expression of miR-19a-3p in each stage of HCC patients was higher than in normal subjects (p < 0.05; Figure 1E). Prognostic analysis of patients with aberrant expression of miR-19a-3p revealed that patients with high expression of miR-19a-3p had significantly shorter and poorer survival than patients with low expression of miR-19a-3p (p < 0.05; Figure 1F). Analysis of miR-376c-3p revealed the identical results (p < 0.05; Figure 1G–I).
Suppression of miR-19a-3p/miR-376c-3p Inhibits HCC Cell Progression
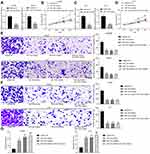
Since the above results proved that miR-19a-3p and miR-376c-3p were highly expressed in HCC tissues and cells, we wondered whether inhibiting miR-19a-3p and miR-376c-3p could retard the malignancy of HCC. In order to verify this conjecture, we transfected Hep3B and Huh7 cells with miR-19a-3p inhibitor or miR-376c-3p inhibitor to silence their expression. The results from Figure 2A and Figure 2C show that miR-19a-3p and miR-376c-3p expression in Hep3B and Huh7 cells was reduced after transfection with miR-19a-3p inhibitor or miR-376c-3p inhibitor, respectively.
The malignant phenotypes of HCC cells were tested by CCK-8 assay, Transwell assay and flow cytometry. Findings demonstrated that Hep3B and Huh7 cells upon treatment of miR-19a-3p inhibitor or miR-376c-3p inhibitor exhibited suppressed proliferation capacity, decreased number of migratory and invasive cells, and elevated apoptosis rate. Moreover, the inhibition of proliferative, migratory, and invasive capacities, together with the promotion of apoptosis rate of Hep3B and Huh7 cells were the most significant when transfected with both miR-19a-3p inhibitor and miR-376c-3p inhibitor (Figure 2B and Figure 2D–G). These findings imply that depleted miR-19a-3p and/or miR-376c-3p can inhibit the malignant phenotypes of HCC cells, and the combined effect is stronger than that of individual miRNA.
SOX6 is a Downstream Target of miR-19a-3p/miR-376c-3p
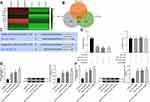
When searching for the targeted genes of miR-19a-3p and miR-376c-3p, we first performed microarray-based analysis on the differentially expressed mRNAs in HCC tissues and adjacent normal tissues, and screened out the mRNAs with poor expression in HCC tissues (Figure 3A). TargetScan database (http://www.targetscan.org/) was then utilized for searching targeted genes of miR-19a-3p and miR-376c-3p, which was intersected with the results of microarray-based analysis (Figure 3B). In the results, we found that both miR-19a-3p and miR-376c-3p had specific binding sites with SOX6 (Figure 3C), suggesting that miR-19a-3p and miR-376c-3p are likely to jointly target SOX6 gene expression.
For verifying this, dual-luciferase reporter gene assay was performed, and the results indicated that miR-19a-3p and miR-376c-3p inhibited SOX6 expression. As shown in Figure 3D, both miR-19a-3p mimic and miR-376c-3p mimic suppressed the expression of SOX6-WT luciferase reporter gene (both p < 0.05), but miR-19a-3p mimic and miR-376c-3p mimic failed to affect SOX6-MUT luciferase reporter gene expression (both p > 0.05). This shows that both miR-19a-3p and miR-376c-3p can inhibit SOX6 expression by targeting the 3ʹUTR of SOX6.
After transfecting miR-19a-3p inhibitor and miR-376c-3p inhibitor into Hep3B and Huh7 cells, SOX6 expression was detected by RT-qPCR and Western blot analysis. The results revealed that cells with miR-19a-3p inhibitor, miR-376c-3p inhibitor, or combination of miR-19a-3p inhibitor and miR-376c-3p inhibitor all elevated SOX6 expression (Figure 3E and F).
SOX6 is Poorly Expressed in HCC Tissues and Cells
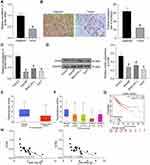
SOX6 mRNA expression in 21 cases of HCC tissues and adjacent normal tissues was tested by RT-qPCR, and the corresponding results revealed that SOX6 mRNA expression in HCC tissues was reduced in contrast to adjacent normal tissues (p < 0.05; Figure 4A). SOX6 positive expression in HCC tissues was detected by immunohistochemical staining, and SOX6 positive rate in HCC tissues was decreased versus adjacent normal tissues (p < 0.05; Figure 4B).
In addition, RT-qPCR and Western blot were also implemented for the detection of SOX6 expression in human normal liver cell line THLE3 and HCC cell lines (Hep3B, MHCC-97L, and Huh7). Similarly, SOX6 expression in HCC cell lines was reduced versus THLE3 cell line (p < 0.05; Figure 4C and D). Detection of the expression of SOX6 in the TCGA database revealed that SOX6 was lowly expressed in HCC patients (p < 0.05; Figure 4E) and poorly expressed in HCC patients at different stages relative to normal controls (p < 0.05; Figure 4F). The survival rate of patients harboring high SOX6 expression benefitted from higher survival (p < 0.05; Figure 4G). Correlation analysis of miR-19a-3p and miR-376c-3p with SOX6, respectively, revealed that miR-19a-3p and miR-376c-3p expression were negatively correlated with SOX6 expression (p < 0.01; Figure 4H). These results suggest that SOX6, poorly expressed in HCC tissues and cells, has prognostic value and negatively correlated with both miR-19a-3p and miR-376c-3p expression in HCC.
miR-19a-3p/miR-376c-3p Participate in the Occurrence of HCC Through Inhibition of SOX6
For the purpose of verifying whether miR-19a-3p and miR-376c-3p were involved in the growth of HCC cells by targeting SOX6, we transfected Hep3B and Huh7 cells with oe-SOX6 alone or in the presence of miR-19a-3p mimic or miR-376c-3p mimic. RT-qPCR was implemented to detect miR-19a, miR-376c, and SOX6 expression. The findings indicated that cells introduced with oe-SOX6 exhibited no difference in miR-19a and miR-376c expression but elevated SOX6 mRNA expression. Increased miR-19a-3p expression or miR-376c-3p expression but reduced SOX6 mRNA expression was found in cells introduced with combination of oe-SOX6 and miR-19a-3p mimic or with oe-SOX6 and miR-376c-3p mimic in comparison with the cells treated with combination of oe-SOX6 and mimic NC (Figure 5A).
The effects of SOX6 on the malignant phenotypes of HCC cells were verified. Findings suggested that Hep3B and Huh7 cells upon treatment of oe-SOX6 presented with suppressed proliferation capacity, decreased number of migratory and invasive cells, and elevated apoptosis rate, while the tendency was opposite in cells with combination of oe-SOX6 and miR-19a-3p mimic or with oe-SOX6 and miR-376c-3p mimic versus the cells treated with combination of oe-SOX6 and mimic NC (Figure 5B–E).
Expression of SOX6 and Wnt/β-catenin pathway-related factors (wnt-5a and β-catenin) was tested by Western blot analysis, and the findings indicated (Figure 5F) that increased SOX6 expression and reduced wnt-5a and β-catenin expression were observed in Hep3B and Huh7 cells upon treatment of oe-SOX6, while expression of which was inverse in cells with combination of oe-SOX6 and miR-19a-3p mimic or oe-SOX6 and miR-376c-3p mimic versus the cells introduced with combination of oe-SOX6 and mimic NC.
This indicates that miR-19a-3p and miR-376c-3p activate the Wnt/β-catenin pathway by targeting SOX6, thereby participating in the HCC cell progression.
miR-19a-3p/miR-376c-3p Inhibitor Inhibit Tumorigenesis of HCC Cells in vivo
In order to confirm that miR-19a-3p and miR-376c-3p retarded the HCC occurrence through regulation of SOX6 in vivo, Hep3B cells with miR-19a-3p or miR-376c-3p inhibitor or oe-SOX6 were injected into nude mice. The results demonstrated that the tumor volume and weight were decreased with treatment of miR-19a-3p/miR-376c-3p inhibitor or oe-SOX6 (Figure 6A–C).
Discussion
Even though many advances have been made in HCC tumor diagnostic and therapeutic approaches, little improvements have been achieved in clinical outcome for HCC patients.13 The predictors of HCC recurrence are largely depended on the clinical or morphological features, and novel researches are necessary for finding new prognostic markers.14 In our study, we aimed to clarify the mechanisms of miR-19a-3p and miR-376c-3p in HCC development, and the corresponding conclusion indicated that miR-19a-3p and miR-376c-3p could target SOX6 to promote the malignant phenotypes of HCC cells in a coordinated manner (Figure 7).
Several articles have demonstrated the important roles of miR-19a-3p in HCC growth. A recent study has revealed that miR-19a-3p is highly expressed in HCC specimens, which leads to HCC metastasis and chemoresistance.8 Another study has indicated that miR-19a-3p is overexpressed in HCC tumors in comparison to respective adjacent tissues, and the knockdown of miR-19a-3p hampered HCC proliferation and invasion, yet promoted cell apoptosis.9 To verify the functions of miR-19a-3p in HCC growth, we detected miR-19a-3p expression in HCC tissues and cells and confirmed its functions in HCC malignancy. The obtained results suggested that miR-19a-3p was highly expressed in HCC, and depleted miR-19a-3p retarded the HCC progression. In accordance with our finding, a recent article has shown that miR-19a-3p promotes HCC cell proliferation in vitro and accelerates liver tumor growth in vivo.15 Similarly, the expression and capabilities of miR-376c in HCC have been discussed before. A recent study has stated that miR-376c-3p is overexpressed in hepatitis B virus-related HCC, indicating its oncogenic roles in HCC.16 Functionally, an article has shown that downregulated miR-376c-3p contributes to suppressed HCC cell progression in vitro and restricted tumor growth in nude mice with HCC.10 In line with prior researches, we found that miR-376c-3p was highly expressed in HCC, and depleted miR-376c-3p also blocked HCC progression. Furthermore, the roles of miR-376c-3p in other diseases have also been demonstrated. For example, it is suggested that restored miR-376c increases the gastric cancer cell growth ability, while depleted miR-376c suppresses these phenotypes.17 Furthermore, we confirmed the combined effects of depleted miR-19a-3p and depleted miR-376c-3p could better inhibit the malignant phenotypes of HCC cells than that of the individual miRNA, as evidenced by more pronounced effects on proliferation, migration, invasion, and apoptosis. However, their combined effects in vivo warrant further exploration.
Subsequently, based on microarray-based analysis and TargetScan database prediction, we found that miR-19a-3p and miR-376c-3p could both target SOX6. SOX6 is considered as an anti-tumor gene, which leads to restricted proliferation and tumorigenicity in diverse cancers, including esophageal squamous cell carcinoma and colorectal cancer.18,19 Xie et al have proposed that miR-155, enhances HCC cell tumorigenesis by targeting SOX6.20 Guo et al have revealed that SOX6 is regarded as a prognostic biomarker for HCC patients,13 and upregulated SOX6 has been revealed to reduce HCC cell progression.21 The ability of SOX6 in HCC malignancy in our study conformed to the aforementioned evidences, which demonstrated that overexpressed SOX6 resulted in suppressed HCC cell proliferation and invasion. Targeting Wnt/β-catenin signaling constitutes a new avenue for treatment for HCC.22 In the Wnt/β-catenin pathway, abnormal modulation of the transcription factor, β-catenin plays a vital part in the progression of early events in HCC.23 The next investigation was concentrated on whether miR-19a-3p and miR-376c-3p were involved in HCC progression via the Wnt/β-catenin signaling by targeting SOX6. As a consequence, wnt-5a and β-catenin expression was then determined. SOX6 is able to bind to β-catenin for β-catenin dissociation from transcriptional complex, thereby preventing its nucleus localization.24 It has been suggested that downregulated SOX6 could alleviate the inhibition of the canonical β-catenin branch through elevating proteasomal degradation of β-catenin.25 Furthermore, the interaction between miR-19a-3p/miR-376c and the Wnt/β-catenin pathway has been highlighted in colorectal cancer and osteoblastogenesis.26,27 Altogether, this article indicated that miR-19a-3p and miR-376c-3p participate in HCC growth by targeting SOX6 and modulating the Wnt/β-catenin pathway.
Conclusion
To conclude, this present study could provide a basis for future research, as miR-19a-3p and miR-376c-3p may have latent capacity as molecular makers for HCC, which could be helpful for HCC therapy. Further discussion of the crosstalk of miRNAs, target gene and their-related pathways may result in future breakthroughs in HCC treatment.
Funding
There is no funding source to report.
Disclosure
All authors declare that there is no conflict of interests in this study.
References
1. Chedid MF, Kruel CRP, Pinto MA, et al. Hepatocellular carcinoma: diagnosis and operative management. Arq Bras Cir Dig. 2017;30(4):272–278. doi:10.1590/0102-6720201700040011
2. Rastogi A. Changing role of histopathology in the diagnosis and management of hepatocellular carcinoma. World J Gastroenterol. 2018;24(35):4000–4013. doi:10.3748/wjg.v24.i35.4000
3. Wang Z, Shi Z, Zhang L, et al. Profilin 1, negatively regulated by microRNA-19a-3p, serves as a tumor suppressor in human hepatocellular carcinoma. Pathol Res Pract. 2019;215(3):499–505. doi:10.1016/j.prp.2018.12.012
4. Zhang Y, Guo X, Li Z, et al. A systematic investigation based on microRNA-mediated gene regulatory network reveals that dysregulation of microRNA-19a/Cyclin D1 axis confers an oncogenic potential and a worse prognosis in human hepatocellular carcinoma. RNA Biol. 2015;12(6):643–657. doi:10.1080/15476286.2015.1022702
5. Qin L, Huang J, Wang G, et al. Integrated analysis of clinical significance and functional involvement of microRNAs in hepatocellular carcinoma. J Cell Physiol. 2019;234(12):23581–23595. doi:10.1002/jcp.28927
6. An Y, Gao S, Zhao WC, et al. Novel serum microRNAs panel on the diagnostic and prognostic implications of hepatocellular carcinoma. World J Gastroenterol. 2018;24(24):2596–2604. doi:10.3748/wjg.v24.i24.2596
7. Lin Q, Chen T, Lin Q, et al. Serum miR-19a expression correlates with worse prognosis of patients with non-small cell lung cancer. J Surg Oncol. 2013;107(7):767–771. doi:10.1002/jso.23312
8. Jiang XM, Yu XN, Liu TT, et al. microRNA-19a-3p promotes tumor metastasis and chemoresistance through the PTEN/Akt pathway in hepatocellular carcinoma. Biomed Pharmacother. 2018;105:1147–1154. doi:10.1016/j.biopha.2018.06.097
9. Qian YY, Liu ZS, Zhang Z, et al. Pterostilbene increases PTEN expression through the targeted downregulation of microRNA-19a in hepatocellular carcinoma. Mol Med Rep. 2018;17(4):5193–5201. doi:10.3892/mmr.2018.8515
10. Wang Y, Chang W, Chang W, et al. MicroRNA-376c-3p facilitates human hepatocellular carcinoma progression via repressing AT-rich interaction domain 2. J Cancer. 2018;9(22):4187–4196. doi:10.7150/jca.27939
11. Sanchez-Mejias A, Kwon J, Chew XH, et al. A novel SOCS5/miR-18/miR-25 axis promotes tumorigenesis in liver cancer. Int J Cancer. 2019;144(2):311–321. doi:10.1002/ijc.31857
12. Hagiwara N. Sox6, jack of all trades: a versatile regulatory protein in vertebrate development. Dev Dyn. 2011;240(6):1311–1321. doi:10.1002/dvdy.22639
13. Guo X, Yang M, Gu H, et al. Decreased expression of SOX6 confers a poor prognosis in hepatocellular carcinoma. Cancer Epidemiol. 2013;37(5):732–736. doi:10.1016/j.canep.2013.05.002
14. Fittipaldi S, Vasuri F, Bonora S, et al. miRNA signature of hepatocellular carcinoma vascularization: how the controls can influence the signature. Dig Dis Sci. 2017;62(9):2397–2407. doi:10.1007/s10620-017-4654-3
15. Sun HX, Yang ZF, Tang WG, et al. MicroRNA-19a-3p regulates cell growth through modulation of the PIK3IP1-AKT pathway in hepatocellular carcinoma. J Cancer. 2020;11(9):2476–2484. doi:10.7150/jca.37748
16. Wang G, Dong F, Xu Z, et al. MicroRNA profile in HBV-induced infection and hepatocellular carcinoma. BMC Cancer. 2017;17(1):805. doi:10.1186/s12885-017-3816-1
17. Hung PS, Chen CY, Chen WT, et al. miR-376c promotes carcinogenesis and serves as a plasma marker for gastric carcinoma. PLoS One. 2017;12(5):e0177346. doi:10.1371/journal.pone.0177346
18. Li H, Zheng D, Zhang B, et al. Mir-208 promotes cell proliferation by repressing SOX6 expression in human esophageal squamous cell carcinoma. J Transl Med. 2014;12(1):196. doi:10.1186/1479-5876-12-196
19. Li YC, Li CF, Chen LB, et al. MicroRNA-766 targeting regulation of SOX6 expression promoted cell proliferation of human colorectal cancer. Onco Targets Ther. 2015;8:2981–2988. doi:10.2147/OTT.S89459
20. Xie Q, Chen X, Lu F, et al. Aberrant expression of microRNA 155 may accelerate cell proliferation by targeting sex-determining region Y box 6 in hepatocellular carcinoma. Cancer. 2012;118(9):2431–2442. doi:10.1002/cncr.26566
21. Li Z, Wang Y. miR-96 targets SOX6 and promotes proliferation, migration, and invasion of hepatocellular carcinoma. Biochem Cell Biol. 2018;96(3):365–371. doi:10.1139/bcb-2017-0183
22. Wang W, Smits R, Hao H, He C. Wnt/beta-catenin signaling in liver cancers. Cancers (Basel). 2019;11(7).
23. Dimri M, Satyanarayana A. Molecular signaling pathways and therapeutic targets in hepatocellular carcinoma. Cancers (Basel). 2020;12(2):491. doi:10.3390/cancers12020491
24. Lin M, Lei T, Zheng J, et al. UBE2S mediates tumor progression via SOX6/beta-catenin signaling in endometrial cancer. Int J Biochem Cell Biol. 2019;109:17–22. doi:10.1016/j.biocel.2019.01.014
25. Leow SC, Poschmann J, Too PG, et al. The transcription factor SOX6 contributes to the developmental origins of obesity by promoting adipogenesis. Development. 2016;143(6):950–961. doi:10.1242/dev.131573
26. Kureel J, John AA, Prakash R, Singh D. MiR 376c inhibits osteoblastogenesis by targeting Wnt3 and ARF-GEF-1 -facilitated augmentation of beta-catenin transactivation. J Cell Biochem. 2018;119(4):3293–3303. doi:10.1002/jcb.26490
27. Yu FB, Sheng J, Yu JM, et al. MiR-19a-3p regulates the forkhead box F2-mediated Wnt/beta-catenin signaling pathway and affects the biological functions of colorectal cancer cells. World J Gastroenterol. 2020;26(6):627–644. doi:10.3748/wjg.v26.i6.627
 © 2021 The Author(s). This work is published and licensed by Dove Medical Press Limited. The full terms of this license are available at https://www.dovepress.com/terms.php and incorporate the Creative Commons Attribution - Non Commercial (unported, v3.0) License.
By accessing the work you hereby accept the Terms. Non-commercial uses of the work are permitted without any further permission from Dove Medical Press Limited, provided the work is properly attributed. For permission for commercial use of this work, please see paragraphs 4.2 and 5 of our Terms.
© 2021 The Author(s). This work is published and licensed by Dove Medical Press Limited. The full terms of this license are available at https://www.dovepress.com/terms.php and incorporate the Creative Commons Attribution - Non Commercial (unported, v3.0) License.
By accessing the work you hereby accept the Terms. Non-commercial uses of the work are permitted without any further permission from Dove Medical Press Limited, provided the work is properly attributed. For permission for commercial use of this work, please see paragraphs 4.2 and 5 of our Terms.