Back to Journals » Cancer Management and Research » Volume 11
MicroRNA-193a-3p suppresses the colorectal cancer cell proliferation and progression through downregulating the PLAU expression
Authors Lin M, Zhang Z , Gao M, Yu H , Sheng H , Huang J
Received 10 March 2019
Accepted for publication 21 May 2019
Published 12 June 2019 Volume 2019:11 Pages 5353—5363
DOI https://doi.org/10.2147/CMAR.S208233
Checked for plagiarism Yes
Review by Single anonymous peer review
Peer reviewer comments 2
Editor who approved publication: Dr Antonella D'Anneo
Maosong Lin,1 Zan Zhang,1 Mingjun Gao,1 Hong Yu,2 Haihui Sheng,3 Junxing Huang4
1Department of Gastroenterology, Taizhou People’s Hospital, Taizhou, Jiangsu 225300, People’s Republic of China; 2Department of Pathology, Taizhou People’s Hospital, Taizhou, Jiangsu 225300, People’s Republic of China; 3Shanghai Engineering Center of Molecular Medicine, and National Engineering Center for Biochip, Shanghai 201203, People’s Republic of China; 4Department of Oncology, Taizhou People’s Hospital, Taizhou, Jiangsu 225300, People’s Republic of China
Background: Colorectal cancer (CRC) is one of the leading causes of cancer-related death in China. Dysregulation of microRNAs (miRNAs) is involved in cancer development and progression. Our previous study showed an inverse relationship between miR-193a-3p expression and the prognosis of CRC. However, the exact biological functions of miR-193a-3p in CRC are still poorly understood. This study aimed to explore the role and mechanism of miR-193a-3p in CRC.
Methods: Real-time PCR and Western blotting were used to examine the expression levels of RNA and protein, respectively. A dual luciferase assay was performed to validate predicted targets of miR-193a-3p. Loss and gain-of-function studies were carried out to reveal the effects and potential mechanism of the miR-193a-3p in the proliferation, metastasis and angiogenesis of CRC cells.
Results: The expression levels of miR-193a-3p in human CRC cell lines were significantly decreased compared with that in normal colonic epithelium cell line. Furthermore, plasminogen activator urokinase (PLAU) was validated as a direct target gene of miR-193a-3p. Over-expression of miR-193a-3p inhibited proliferation, migration and angiogenesis of HT-29 cell, whereas forced expression of PLAU could rescue the inhibitory effects.
Conclusion: miR-193a-3p might inhibit CRC cell growth, migration and angiogenesis partly through targeting PLAU. MiR-193a-3p/PLAU axis might provide a potent therapeutic opportunity for aggressive CRC.
Keywords: microRNA-193a-3p, colorectal cancer, PLAU, cell proliferation, invasion, angiogenesis
Introduction
Recently, there is a pleasant change of incidence and mortality in some gastrointestinal (GI) malignant tumors such as gastric, esophageal and liver cancers in both sexes in China. However, colorectal cancer (CRC),the third most common cancer worldwide, did not show same parallel trend and manifested the opposite direction.1 From 2000 to 2011, a significant upward tendency in age-standardized incidence and mortality rates of CRC was observed especially in men, which ranked NO.1.2 Despite intensive efforts being made, the improvement of survival rate of CRC patients is still limited, and our knowledge of the molecular mechanisms that lead to the development and progression of CRC remains largely unknown.3,4 Therefore, it is an urgent need to elucidate the exact intrinsic mechanism of the tumorgenesis of CRC including genetic and epigenetic alterations and then innovative early diagnosis and treatment algorithms.
MicroRNAs (miRNAs), a class of small non-coding RNA molecules, play critical roles in a variety of biological events, including development, cell proliferation and differentiation through regulating their target genes expression by binding to the 3′-un-translated region (UTR).5,6 As a result, it is not surprising that miRNAs can widely involve in human diseases. Emerging evidence has now demonstrated that aberrantly expressed miRNAs have critical implications with regard to the initiation and progression of various types of cancers.7,8,10,14 Dysregulation of miR-193a-3p, a gene located on human chromosome 17q11.2 (chr17:31,558,803–31,560,358), has been found involved in several cancers including CRC.9–11 Besides the oncogenic role being reported only in a relatively very few studies,12 almost the vast majority of published data pointed toward a role of miR-193a-3p as tumor suppressor in both solid and blood cancers by targeting targets with potential oncogenic functions to act its tumor cell aggressive inhibiting properties.13–17 This contradiction phenomenon also could be seen in studies focused on CRC.18–20 Overexpression of miR-193a-3p inhibited colon cancer cell proliferation and induce apoptosis.19 However, Yong et al18 reported that miR-193a-3p was upregulated in both tissue and blood samples and its expression level was increased with disease progression. The role of miR-193a-3p in CRC may be dependent on the cellular context. Our previous study revealed that miR-193a-3p was downregulated in CRC tissues and was associated with the prognosis of CRC patients.21 Therefore, the exact underlying molecular mechanisms of miR-193a-3p in CRC are still need well characterized.
Plasminogen activator urokinase (PLAU), also known as urokinase plasminogen activator (uPA), codes a serine protease which can bind to its receptor and then promote a proteolytic cascade to convert these proteases into their active forms which can confer tumor cells with the ability to degrade the components of the surrounding extracellular matrix (ECM).22–24 As a result, PLAU can involve in tumor cell migration, invasion as well as metastasis as a key player.25,26 However, the biological role of PLAU and its inter mechanisms in CRC remain unclear.
In this study, miR-193a-3p was found downregulated in CRC cells and forced over-expression of miR-193a-3p inhibited HT-29 cells proliferation, migration as well as angiogenesis. Moreover, PLAU was identified as a directed target of miR-193a-3p. Taken together, we suggested that miR-193a-3p might serve as a tumor suppressor in CRC and miR-193a-3p/PLAU axis might be a potential candidate for the treatment of CRC.
Materials and methods
Cell lines and cell culture
Three CRC cell lines including HT29,HCT116 and LOVO, one normal colonic epithelium cell line NCM460 and human umbilical vein endothelial cells (HUVEC) were purchased from the Cell Bank of Type Culture Collection of Chinese Academy of Sciences (Shanghai, China). The HT29 and LOVO cells were cultured in Dulbecco’s modified Eagle’s medium (DMEM; Invitrogen, CA, USA), while HCT116 and NCM460 cells were cultured in 1640 medium supplemented with 10% fetal bovine serum (FBS; Invitrogen, CA, USA) and 1% glutamine at 37°C with 5% CO2.
Total RNA isolation and quantitative real-time PCR (qPCR)
Total RNAs of three CRC cells and NCM460 cells were isolated using TRIzol reagent respectively (Takara, Tokyo, Japan) according to the manufacturer’s instructions. For miR-193a-3p, reverse transcription reactions were performed with an All-in-One™ miRNA qRT-PCR Detection Kit (GeneCopoeia, MD, USA) to obtain the complementary DNA (cDNA) templates. The qPCR was performed in an iQ5 Real-Time PCR Detection Systems (Bio-Rad, CA, USA) using All-in-One™ miRNA qRT-PCR Detection Kit (GeneCopoeia, MD, USA). The relative expression levels of miR-193a-3p were normalized to the expression level of human actin beta (hACTB). The PCR primes for miR-193a-3p and hACTB are as follows: miR-193a-3p: forward: 5ʹ-AACTGGCCTACAAAGTCCCAGT-3, and the reverse primer was provided in the All-in-One™ miRNA qRT-PCR Detection Kit. hACTB: forward: 5′GGCACTCTTCCAGCCTTCC-3′, reverse:5′-GAGCCGCCGATCCACAC-3′. The data analyses were performed using the 2−ΔΔCt method.
miRNA mimics, inhibitor, PLAU plasmid and siRNA transfection
MiR-193a-3p mimics, inhibitor and negative control (NC) were purchased from GenePharma (Shanghai, China) and pcDNA3.1-PLAU, and blank vector were purchased from Genecreat (Wuhan, China). The siRNA sequence targeting the human PLAU cDNA was designed and synthesized by GenePharma (Shanghai, China). The siRNA sequence was 5′-CGGGAAUCUCAUCUUUCUU-3′. HT29 cells were seeded in 12-well plates. After 12 hrs, cells were transiently transfected by using Lipofectamine 2000 (Thermo Fisher Scientific, MA, USA) according to the manufacturer’s instructions.
Dual luciferase assay
TargetScan (
The cancer genome atlas (TCGA) database bioinformatics analysis
The Gene Expression Profiling Interactive Analysis (GEPIA) database was used to examine PLAU expression in CRC.27 A total of 275 colon adenocarcinoma (COAD) and 349 normal tissues data obtained from the TCGA database were enrolled in this study to examine whether any significant differences in PLAU expression existed between normal and colon cancer tissues. Fold change >1.5 and P-value <0.05 between the tumor and normal tissues were considered as significant.
Cell proliferation assays
The effect of miR-193a-3p on growth of HT-29 cell line was carried out as previously described. Briefly, HT-29 cells (5×103/well) were seeded into a 96-well plate at a density of 5×103/well in triplicate. At 1, 2, 3, 4 and 5 days after incubation, cell proliferation was assessed by using cell counting kit-8 assay (CCK-8; Dojindo, Kumamoto, Japan) according to the manufacturer’s instructions. Absorbance was measured at 450 nm using a MultiskanMK3 microplate reader (Thermo Fisher Scientific, MA, USA).
Transwell assay
To detect the migratory capability, 8×105 HT-29 cells/well were seeded into a transwell apparatus (Corning, NY, USA) with the serum-free medium. Relatively, medium containing 10% serum functioned as a chemoattractant in the lower chamber. After incubation for 48 hrs, the stained cells were photographed and counted by a microscope.
Tube formation assay
HUVECs (4×104 cells) were suspended in culture supernatants of HT-29 cells and DMEM containing 10% FBS (100 µL), and then seeded into a 96-well plate precoated with 50 µL/well Matrigel Basement Membrane Matrix (BD Biosciences, NJ, USA). HUVECs were cultured in 100 µL DMEM at 37°C for 6 hrs. Images were taken using a bright-field microscope.
Western blotting
Briefly, cell pallets of each group were lysed in the RIPA buffer containing protease inhibitors and phosphatase inhibitors (Sigma, MO, USA). Proteins were extracted according to the manufacturer’s instructions, and the concentration was determined by the Bradford assay. Thirty micrograms protein samples were separated on a sodium dodecylsulfate-polyacrylamide gelelectrophoresis(SDS-PAGE) and transferred to a 0.2 μm polyvinylidene difluoride membrane. The membrane was blocked in 5% non-fat milk. Membranes were incubated with primary antibodies against PLAU (1:1,000, Abcam, CA, USA) and tubulin (1:5,000, Abcam, CA, USA) overnight at 4°C. After washed 3 times with Tris-buffered saline (TBST), membranes were incubated with horseradish peroxidase-conjugatedanti-mouse or anti-rabbit IgG (1:5,000, Santa Cruz Biotechnology, CA, USA) for 2 hrs at room temperature, and then washed 3 times with TBST. The signal was visualized by an enhanced chemiluminescence solution (Amersham Pharmacia Biotech, Uppsala, Sweden).
Statistical analyses
All experiments were performed in triplicate, and statistical analyses were performed using SPSS 19.0 software (SPSS Inc., IL, USA). Data were presented as the means and error bars indicated the standard deviation (SD). A Student’s t-test was used for the statistical comparison of data. One-way analysis of variance was used to compare differences between the means of three groups. Statistical differences were considered significantly if a P-value was less than 0.05.
Results
miR-193a-3p is downregulated in CRC cell lines
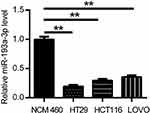
To confirm the expression of miR-193a-3p in CRC cells, qPCR was used to detect the expression levels of miR-193a-3p in HT29, HCT116 and LOVO cell lines as well as normal colonic epithelial cell line NCM460. The results showed that miR-193a-3p was significantly decreased in all CRC cells in comparison to NCM460 cells (Figure 1, P<0.01). HT29 cells had the lowest expression levels of miR-193a-3p and then was selected for further experiments.
miR-193a-3p inhibits the proliferation, migration and angiogenesis of HT-29 cells
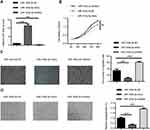
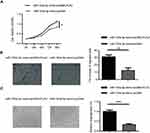
To determine the role of miR-193a-3p in regulating the progression of CRC, we performed gain and loss-of-function experiments, using miR-193a-3p mimics and inhibitor, respectively. The qPCR assays showed that miR-193a-3p was significantly upregulated or downregulated in HT29 cells transfected with miR-193a-3p mimics or inhibitor, respectively, in comparison to negative control transfection (Figure 2A, P<0.001 or P<0.01). CCK-8 assay was used to evaluate the effect of miR-193a-3p on the proliferation of HT29 cells. The data revealed that the proliferation rate was dramatically decreased in the miR-193a-3p over-expression cells and increased in the miR-193a-3p knockdown cells compared with the control group (Figure 2B, P<0.01).
We further performed transwell assay to assess the effect of miR-193a-3p on migration in HT29 cells. As shown in Figure 2C, over-expression of miR-193a-3p significantly inhibited the migration ability of HT29 cells (P<0.001), while knockdown of miR-193a-3p promoted migration of HT-29 cells compared with the control group (P<0.001). Taken together, these data indicated that miR-193a-3p negatively regulates CRC cell proliferation and migration in vitro.
In addition, the effect of miR-193a-3p on angiogenic activity was evaluated using tube formation assay. As shown in Figure 2D, over-expression of miR-193a-3p significantly inhibited tube-like structure formation of HUVECs (P<0.001), while knockdown of miR-193a-3p led to promoting inhibited tube-like structure formation of HUVECs compared with the control group (P<0.001). This result indicates that miR-193a-3p suppresses the angiogenic activity of CRC cells.
PLAU is a direct target gene of miR-193a-3p
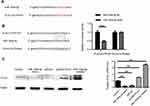
To clarify the underlying molecular mechanisms of miR-193a-3p in CRC, three miRNA target-prediction website TargetScan, picTar and miRnada were used to predict targets of miR-193a-3p. We found that PLAU was predicted as a potential target of miR-193a-3p. The 3ʹ-UTR of PLAU has a binding site for miR-193a-3p (Figure 3A). Dual luciferase assay showed that the luciferase activity of wild-type 3ʹ-UTR of PLAU was inhibited by miR-193a-3p, while that of mutant 3ʹ-UTR did not exhibit the same effect (Figure 3B, P<0.001). These findings imply that miR-193a-3p may directly bind to the 3ʹ-UTR of PLAU and downregulate its expression at a post-transcriptional level. Subsequent Western blot analysis verified that the protein levels of PLAU were relatively higher in HT29 cells transfected with inhibitor and lower with mimic than that in control cells (Figure 3C, P<0.01). Based on the above-mentioned results, we could conclude that PLAU was a direct target gene of miR-193a-3p in CRC cells.
PLAU enhances the proliferation, migration and angiogenic abilities of HT-29 cells
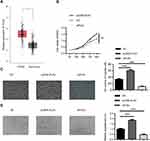
To explore the role of PLAU in the progression of CRC, we firstly analyzed the expression profiles data from TCGA database. The results revealed that the expression levels of PLAU in 274 colon adenocarcinoma were higher than those in 349 normal tissues (Figure 4A, P<0.05). Then, HT29 cells were transfected with pcDNA-PLAU encoding the full-length PLAU and siPLAU. The transfection and expression efficiency were confirmed by Western blot (Figure 3C). The ability of cell proliferation, migration and angiogenesis were assessed. CCK-8 assay showed that over-expression of PLAU significantly promoted cell proliferation (Figure 4B, P<0.01), while knockdown of PLAU significantly inhibited cell proliferation compared with the inhibited control group (Figure 4B, P<0.01). In addition, the migration capacity of HT-29 cells was significantly enhanced by overexpression of PLAU (Figure 4C, P<0.001). In contrast, the migration capacity of HT29 cells was remarkably suppressed by knockdown of PLAU (Figure 4C, P<0.001). Tube formation assay revealed that over-expression of PLAU significantly facilitated tube-like structure formation (Figure 4D, P<0.001), while knockdown of PLAU significantly suppressed tube-like structure formation compared with the inhibited control group (Figure 4D, P<0.001).
PLAU is involved in the miR-193a-3p induced suppression of CRC cell growth, migration and angiogenesis
To further explore the functional connection between miR-193a-3p and PLAU, HT29 cells were co-transfected with miR-193a-3p mimic and PLAU plasmid or blank vector. The suppressive effect of miR-193a-3p on cell proliferation rate could be abolished after co-transfected with miR-193a-3p mimics and PLAU plasmid (Figure 5A, P<0.05). Besides, the results of transwell assay showed that miR-193a-3p over-expression significantly reduced the ability of cell migration, whereas cells co-transfected with miR-193a-3 mimics and PLAU plasmid could counteract the suppressive effect (Figure 5B, P<0.01). Moreover, few previous studies have linked the tumor suppressor miR-193a-3p to changes in cell angiogenesis. In the present study, the tube formation assays results manifested that overexpression of PLAU was further shown to reverse the inhibitory effects of miR-193a-3p on cell angiogenesis CRC (Figure 5C, P<0.001).
Taken together, these results suggested that the effects of miR-193a-3p appear to be due, at least in part, to downregulation of PLAU.
Discussion
Increasing evidences have indicated the pivotal role of miR-193a-3p in the initiation and progression of several cancers including CRC.14–16 However, to the best of our knowledge, the underlying mechanisms of miR-193a-3p in CRC development are still poorly understood.
Two studies from well-regarded groups reported that miR-193a-3p was usually under-expressed in CRC cells and acting as a tumor suppressor.17,20 Our previous report extended their finding by identifying that decreased expression of miR-193a-3p was associated with poor prognosis in CRC.21
In the current report, we intended to use established CRC cell lines to functionally explore the role of miR-193a-3p in CRC metastasis. We initially confirmed that miR-193a-3p was significantly downregulated in all three CRC cell lines analyzed.
Moreover, we found that forced over-expression of miR-193a-3p not only significantly reduced the CRC cells’ proliferation but also decreased their abilities of migration and angiogenesis. These results suggested the tumor suppressor role of miR-193a-3p in CRC.
It has been widely confirmed that miRNAs exert their functions by binding to their target genes. Previous researches have shown that miR-193a-3p could serve as a tumor suppressor via targeting several genes including cyclinD1, KRAS,as well as IL17RD and so on involved in diverse tumors.13,16,19 Actually, for hsa-miR-193a-3p, bioinformatics approaches predicted 293 putative target genes with 168 displaying the highest score (cumulative weighted context++ score <−0.24). Among them, PLAU contains a perfect binding sequence for miR-193a-3p in the 3ʹ UTR. To support this bioinformatics prediction, a recent study by Iwamoto et al reported downregulation of miR-193b in skin biopsies and fibroblasts from patients with systemic sclerosis and this downregulation was correlated with urokinase-type plasminogen activator upregulation.28 To confirm PLAU as the direct target of miR-193a-3p in CRC, we performed targeted gene analysis by cloning PLAU 3ʹ UTR sequence with predicted binding to miR-193a-3p into luciferase reporter vector and coexpressed with miR-193a-3p vector in CRC HT29 cells. Co-expression of miR-193a-3p significantly reduced reporter gene expression, indicating PLAU 3ʹ UTR sequence is a direct target of miR-193a-3p in CRC cells. To further support the results from our reporter gene analysis, we induced forced expression of miR-193a-3p in HT29 cells via transient transfection. Significant reduction of PLAU protein was found in the HT29 cells 48 hrs after transient expression of miR-193a-3p.
It is worthy to point out that although our work is not the first to indicate PLAU as the miR-193a-3p target overall, we indeed provided direct evidence to demonstrate that miR-193a-3p can directly downregulate PLAU expression via a miR-193a-3p binding target in PLAU 3ʹ untranslated region.
PLAU, a serine proteinase secreted by many cancer cells including CRC cells, can bind to the urokinase-type plasminogen activator receptor (uPAR) and transform inactive plasmin and other proteases, including matrix metallopeptidase 9 (MMP9), into their active forms and then to stimulate tumor cell proliferation and adhesion.24,29 In particular, PLAU can accelerate tumor metastasis and promote tumor angiogenesis by degrading ECM and basement membranes such as laminin, fibronectin and collagen, allowing cells to migrate.23,30 It has been reported that the high level of PLAU indicates poor prognosis, and American Society Clinical Oncology (ASCO) has appealed that PLAU should act as risk assessment and a possible treatment target.31 The important role of PLAU in cell proliferation and tumorigenesis has been well characterized. Tang et al found that downregulated uPA could cause notable decrease for the complete channels formed by SKOV-3 and OVCAR-3 cells. Their mechanism study further disclosed uPA could promote vasculogenic minicry (VM) formation through regulating AKT/mTOR/MMP-2/Laminin5γ2 signal pathway. They argued that PLAU might serve as therapeutic target of VM for ovarian cancer.32 Moirangthem et al confirmed that simultaneous silencing of uPA and MMP9 in breast cancer cells decreased the wound healing, migratory, invasive and adhesive capacity of the cells. A remarkable increase in the expression of cell-to-cell adhesion molecule E-cadherin and decrease in Vimentin and Snail expression were also detected.26 In the present study, through the TCGA analysis, PLAU was found over-expressed in CRC tissues in comparison to the normal controls, which indicates the functional oncogenic role of PLAU in the progression of CRC. Furthermore, we found the commonly upregulated PLAU expression through transfect with PLAU plasmid in HT29 cells could accelerate the cell proliferation, migration as well as angiogenesis potent while downregulation of this gene by using siRNA transfection could get reverse effect, which confirmed its tumorgenesis role in CRC. Previous study speculated that the interaction between uPA and uPAR results in the release of signaling molecules that stimulate cell proliferation/survival and tumor development.33 As a result, we could suggest that PLAU might involve in the tumor growth and metastasis in CRC. However, the exact downstream pathways need more investigation.
Although miR-193a-3p was found as a negative regulator of PLAU in other human cancers and the high expression of miR-193a could strongly inhibit PLAU expression and decreased cell aggressive properties, relatively fewer studies focused on this topic in CRC was now found.34–36 Thereafter, using a series of in vitro assays, we further investigated whether PLAU mediated the suppressive ability of miR-193a-3p in CRC cell. Encouragingly, our findings exhibited that over-expression of miR-193a-3p could reduce PLAU at protein levels, and inhibition of PLAU expression could simulate the suppressing effect of miR-193a-3p mimic in HT-29 cells while upregulation of PLAU by transfecting with PLAU plasmid could reverse the decrease of proliferation, invasion and angiogenesis caused by miR-193a-3p mimic in vitro. Therefore, the present results might provide a novel and comprehensive insight into the functional role of miR-193a-3p in CRC by directly targeting PLAU.
Our understanding of the downstream of PLAU to drive tumor cell migration, metastasis and angiogenesis has advanced considerably in the last ten years, but our understanding of what occurs upstream of PLAU modulated mechanisms remains relatively limited. There have been findings which suggest that demethylation of the uPA promoter can be the mechanism responsible for induction of uPA expression in the initiation of early stages of breast cancer.37 In our study, forced over-expression of PLAU in HT-29 cells could promote the cell capacity of proliferation, migration as well as angiogenesis. Moreover, using a series of in vitro assays, we also found the expression of PLAU could be regulated by miR-193a-3p and then PLAU could mediate the suppressive ability of miR-193a-3p in this CRC cell. Therefore, we argued that the promoting effects of miRNA deregulation might be a novel pivotal regulatory mechanism responsible for the induction of PLAU expression during CRC tumorigenesis.
In conclusion, our study manifested that miR-193a-3p expression was decreased in CRC cell lines, and upregulation of miR-193a-3p inhibited tumor development and progression in vitro through regulating cell growth, migration and angiogenesis partly through targeting the PLAU pathway. Although the exact role of miR-193a-3p/PLAU pathway in CRC needs furthermore investigated, these observations actually provided new evidence for targeting this newly identified regulatory signaling pathway which provides therapeutic opportunities for aggressive CRC.
Acknowledgments
The present study was supported by the Project of Health Department of Jiangsu Province, China (grant no. H201363), the Social Development Project of Taizhou City, Jiangsu, China (grant no. TS025), the Project of Taizhou People’s Hospital (grant no. 2016) and Jiangsu Provincial Medical Innovation Team (grant no. CXTDA2017042).
Disclosure
The authors report no conflicts of interest in this work.
References
1. Chen W, Zheng R, Baade PD, et al. Cancer statistics in China, 2015. CA Cancer J Clin. 2016;66(2):115–132. doi:10.3322/caac.21338
2. Yu W, Jiang J, Xie L, et al. Mortality trends in colorectal cancer in China during 2000–2015: a joinpoint regression and age–period cohort analysis. Prev Chronic Dis. 2018;15:E156. doi:10.5888/pcd15.180329
3. Gill S, Blackstock AW, Goldberg RM. Colorectal cancer. Mayo Clin Proc. 2007;82:114–129. doi:10.1016/S0025-6196(11)60974-9
4. Siegel R, Desantis C, Jemal A. Colorectal cancer statistics, 2014. CA Cancer J Clin. 2014;64:104–117. doi:10.3322/caac.21220
5. Bertoli G, Cava C, Castiglioni I. MicroRNAs: new biomarkers for diagnosis, prognosis, therapy prediction and therapeutic tools for breast cancer. Theranostics. 2015;5(10):1122–1143. doi:10.7150/thno.11543
6. Choi E, Hwang KC. MicroRNAs as novel regulators of stem cell fate. World J Stem Cells. 2013;5:172–187. doi:10.4252/wjsc.v5.i4.172
7. Zhang G, Zhou H, Xiao H, Liu Z, Tian H, Zhou T. MicroRNA-92a functions as an oncogene in colorectal cancer by targeting PTEN. Dig Dis Sci. 2014;59:98–107. doi:10.1007/s10620-013-2858-8
8. Ling H, Pickard K, Ivan C, et al. The clinical and biological significance of miR-224 expression in colorectal cancer metastasis. Gut. 2016;65(6):977–989. doi:10.1136/gutjnl-2015-309372
9. Kent WJ, Sugnet CW, Furey TS, et al. The human genome browser at UCSC. Genome Res. 2002;12(6):996–1006. doi:10.1101/gr.229102
10. Yu T, Li J, Yan M, et al. MicroRNA-193a-3p and −5p suppress the metastasis of human non-small-cell lung cancer by downregulating the ERBB4/PIK3R3/mTOR/S6K2 signaling pathway. Oncogene. 2015;34(4):413–423. doi:10.1038/onc.2014.462
11. Mamoori A, Wahab R, Islam F, et al. Clinical and biological significance of miR-193a-3p targeted KRAS in colorectal cancer pathogenesis. Hum Pathol. 2018;71:145–156. doi:10.1016/j.humpath.2017.10.024
12. Yi Y, Chen J, Jiao C, et al. Upregulated miR-193a-3p as an oncogene in esophageal squamous cell carcinoma regulating cellular proliferation, migration and apoptosis. Oncol Lett. 2016;12(6):4779–4784. doi:10.3892/ol.2016.5229
13. Liu YF, Xu X, Xu XL, et al. MicroRNA-193a-3p inhibits cell proliferation in prostate cancer by targeting cyclin D1. Oncol Lett. 2017;14(5):5121–5128.
14. Pu Y, Zhao F, Cai W, Meng X, Li Y, Cai S. MiR-193a-3p and miR-193a-5p suppress the metastasis of human osteosarcoma cells by down-regulating Rab27B and SRR, respectively. Clin Exp Metastasis. 2016;33:359–372. doi:10.1007/s10585-016-9783-0
15. Williams M, Kirschner MB, Cheng YY, et al. miR-193a-3p is a potential tumor suppressor in malignant pleural mesothelioma. Oncotarget. 2015;6(27):23480–23495. doi:10.18632/oncotarget.4346
16. Pekow J, Meckel K, Dougherty U, et al. miR-193a-3p is a key tumor suppressor in ulcerative colitis-associated colon cancer and promotes carcinogenesis through upregulation of IL17RD. Clin Cancer Res. 2017;23(17):5281–5291. doi:10.1158/1078-0432.CCR-17-0171
17. Takahashi H, Takahashi M, Ohnuma S, et al. microRNA-193a-3p is specifically downregulated and acts as a tumor suppressor in BRAF-mutated colorectal cancer. BMC Cancer. 2017;17(1):723. doi:10.1186/s12885-017-3739-x
18. Yong FL, Law CW, Wang CW. Potentiality of a triple microRNA classifier: miR-193a-3p, miR-23a and miR-338-5p for early detection of colorectal cancer. BMC Cancer. 2013;13:280. doi:10.1186/1471-2407-13-280
19. Seviour EG, Sehgal V, Mishra D, et al. Targeting KRas-dependent tumor growth, circulating tumor cells and metas tasis in vivo by clinically significant miR-193a-3p. Oncogene. 2017;36(10):1339–1350. doi:10.1038/onc.2016.308
20. Teng Y, Ren Y, Hu X, et al. MVP-mediated exosomal sorting of miR-193a promotes colon cancer progression. Nat Commun. 2017;8:14448. doi:10.1038/ncomms14448
21. Lin MS, Duan BS, Hu JF, et al. Decreased expression of miR-193a-3p is associated with poor prognosis in colorectal cancer. Oncol Lett. 2017;14:1061–1067. doi:10.3892/ol.2017.6266
22. Montuori N, Pesapane A, Rossi FW, et al. Urokinase type plasminogen activator receptor (uPAR) as a new therapeutic target in cancer. Transl Med UniSa. 2016;15(3):15–21.
23. Brungs D, Chen J, Aghmesheh M, et al. The urokinase plasminogen activation system in gastroesophageal cancer: a systematic review and meta-analysis. Oncotarget. 2017;8(14):23099–23109. doi:10.18632/oncotarget.15485
24. Mahmood N, Mihalcioiu C, Rabbani SA. Multifaceted role of the urokinase-type plasminogen activator (uPA) and its receptor (uPAR): diagnostic, prognostic, and therapeutic applications. Front Oncol. 2018;12(8):24. doi:10.3389/fonc.2018.00024
25. Mauro CD, Pesapane A, Formisano L, et al. Urokinase-type plasminogen activator receptor (uPAR) expression enhances invasion and metastasis in RAS mutated tumors. Sci Rep. 2017;7(1):9388. doi:10.1038/s41598-017-10062-1
26. Moirangthem A, Bondhopadhyay B, Mukherjee M, et al. Simultaneous knockdown of uPA and MMP9 can reduce breast cancer progression by increasing cell-cell adhesion and modulating EMT genes. Sci Rep. 2016;6:21903. doi:10.1038/srep21903
27. Tang Z, Li C, Kang B, Gao G, Li C, Zhang Z. GEPIA: a web server for cancer and normal gene expression profiling and interactive analyses. Nucleic Acids Res. 2017;45:W98–W102. doi:10.1093/nar/gkx247
28. Iwamoto N, Vettori S, Maurer B, et al. Downregulation of miR-193b in systemic sclerosis regulates the proliferative vasculopathy by urokinase-type plasminogen activator expression. Ann Rheum Dis. 2016;75:303–310. doi:10.1136/annrheumdis-2014-205326
29. Su SC, Lin CW, Yang WE, Fan WL, Yang SF. The urokinase-type plasminogen activator (uPA) system as a biomarker and therapeutic target in human malignancies. Expert Opin Ther Targets. 2016;20(5):551–566. doi:10.1517/14728222.2016.1113260
30. Mekkawy AH, Pourgholami MH, Morris DL. Involvement of urokinase-type plasminogen activator system in cancer: an overview. Med Res Rev. 2014;34(5):918–956.
31. Bellocq JP, Luporsi E, Barriere J, et al. uPA/PAI-1, Oncotype DX, Mammaprint((R)). Prognosis and predictive values for clinical utility in breast cancer management. Ann Pathol. 2014;34(5):349–351. doi:10.1016/j.annpat.2014.04.010
32. Tang J, Wang J, Fan L, et al. cRGD inhibits vasculogenic mimicry formation by down-regulating uPA expression and reducing EMT in ovarian cancer. Oncotarget. 2016;7(17):24050–24062.
33. Chiang SP, Cabrera RM, Segall JE. Tumor cell intravasation. Am J Physiol Cell Physiol. 2016;311(1):C1–C14. doi:10.1152/ajpcell.00238.2015
34. Iliopoulos D, Rotem A, Struhl K. Inhibition of miR-193a expression by Max and RXRα activates K-Ras and PLAU to mediate distinct aspects of cellular transformation. Cancer Res. 2011;71(15):5144–5153. doi:10.1158/0008-5472.CAN-11-0425
35. Salvi A, Conde I, Abeni E, et al. Effects of miR-193a and sorafenib on hepatocellular carcinoma cells. Mol Cancer. 2013;12:162. doi:10.1186/1476-4598-12-162
36. Noh H, Hong S, Dong Z, Pan ZK, Jing Q, Huang S. Impaired MicroRNA processing facilitates breast cancer cell invasion by upregulating urokinase-type plasminogen activator expression. Genes Cancer. 2011;2(2):140–150.
37. Pakneshan P, Teˆtu B, Rabbani SA. Demethylation of urokinase promoter as a prognostic marker in patients with breast carcinoma. Clin Cancer Res. 2004;10(9):3035–3041. doi:10.1158/1078-0432.CCR-04-1002
 © 2019 The Author(s). This work is published and licensed by Dove Medical Press Limited. The full terms of this license are available at https://www.dovepress.com/terms.php and incorporate the Creative Commons Attribution - Non Commercial (unported, v3.0) License.
By accessing the work you hereby accept the Terms. Non-commercial uses of the work are permitted without any further permission from Dove Medical Press Limited, provided the work is properly attributed. For permission for commercial use of this work, please see paragraphs 4.2 and 5 of our Terms.
© 2019 The Author(s). This work is published and licensed by Dove Medical Press Limited. The full terms of this license are available at https://www.dovepress.com/terms.php and incorporate the Creative Commons Attribution - Non Commercial (unported, v3.0) License.
By accessing the work you hereby accept the Terms. Non-commercial uses of the work are permitted without any further permission from Dove Medical Press Limited, provided the work is properly attributed. For permission for commercial use of this work, please see paragraphs 4.2 and 5 of our Terms.