Back to Journals » OncoTargets and Therapy » Volume 12
MicroRNA-186-5p Inhibits Proliferation And Metastasis Of Esophageal Cancer By Mediating HOXA9
Authors Xu C, Li B, Zhao S, Jin B, Jia R, Ge J, Xu H
Received 20 August 2019
Accepted for publication 15 October 2019
Published 30 October 2019 Volume 2019:12 Pages 8905—8914
DOI https://doi.org/10.2147/OTT.S227920
Checked for plagiarism Yes
Review by Single anonymous peer review
Peer reviewer comments 2
Editor who approved publication: Prof. Dr. Takuya Aoki
Changqin Xu, Bin Li, Shulei Zhao, Bingjie Jin, Ruzhen Jia, Jian Ge, Hongwei Xu
Department of Gastroenterology, Shandong Provincial Hospital Affiliated to Shandong University, Jinan, People’s Republic of China
Correspondence: Hongwei Xu
Department of Gastroenterology, Shandong Provincial Hospital Affiliated to Shandong University, 324 Jingwu Road, Huaiyin District, Jinan, Shandong 250021, People’s Republic of China
250021 Email [email protected]
Objective: MicroRNA (miRNA) is an endogenous, non-coding small RNA that plays a key role in regulating organism biology and pathology. The aim of this study was to investigate the expression characteristics of microRNA-186-5p in esophageal cancer (ECa) and its correlation with clinical progression and prognosis, and to further explore its underlying mechanisms.
Methods: Real-time quantitative PCR (qRT-PCR) was used to detect microRNA-186-5p level in 45 pairs of ECa tissue samples and adjacent ones, and to analyze the expression of microRNA-186-5p and clinical progression of ECa and prognosis. The relationship between microRNA-186-5p level in ECa cell lines was further verified by qRT-PCR. Finally, the potential mechanism was explored using luciferase reporter gene assay and cell recovery experiment.
Results: QRT-PCR results revealed that the expression of microRNA-186-5p in ECa tissues was remarkably lower than that in adjacent tissues, and the difference was statistically significant. Compared with patients with high expression of microRNA-186-5p, patients with low expression of microRNA-186-5p had higher incidence of pathological stage and lower overall survival rate. Besides, compared with the miR-NC group, the microRNA-186-5p mimics group had a significant decrease in proliferation and metastasis ability of ECa cells. Subsequent qRT-PCR validation in ECa cell lines and tissues indicated a significant increase in HOXA9 expression and a negative correlation with microRNA-186-5p.
Conclusion: The expression of microRNA-186-5p was remarkably decreased in ECa, which was remarkably correlated with pathological stage, distant metastasis and poor prognosis of ECa. The results suggested that microRNA-186-5p may inhibit cell proliferation of ECa by regulating HOXA9.
Keywords: microRNA-186-5p, HOXA9, esophageal cancer, proliferation, metastasis
Introduction
Esophageal cancer (ECa) is one of the alimentary tract malignancies with high morbidity and mortality. Currently, it ranks 9th in the global cancer incidence and 6th in the total mortality.1–3 In developed countries, the incidence of ECa ranks 20th, but its death ranks 11th.4,5 In developing countries, the incidence of ECa ranks 8th and the mortality ranks 5th.6 Among esophageal carcinoma, squamous cell carcinoma and adenocarcinoma are the main pathological types, among which squamous cell carcinoma accounts for more than 90% in China.7 Because the early symptoms of ECa are not obvious and there is a lack of early diagnosis, most patients are in the advanced stage of disease at the time of the first diagnosis, and the 5-year survival rate of ECa after resection is 9.5–45%.8 Tumor invasion and metastasis are the main factors leading to the death of patients with ECa, which are mainly dependent on the activation of proto-oncogenes and the inactivation of tumor suppressor genes.9,10 If we can study the occurrence and development of ECa from the perspective of genetics and epigenetics, it will provide new targets and new strategies for the diagnosis and treatment of ECa.11,12
MicroRNA (miRNA) are newly-discovered small non-coding RNA molecules with about 22 nucleotides in length, and are widely distributed in eukaryotes.13,14 Although most miRNAs are intracellular, some have been found in the extracellular environment, called circulating miRNAs or extracellular miRNAs.15,16 MiRNAs bind to some proteins to form miRNA-induced silencing complexes, which in turn lead to complete or incomplete pairing with the 3ʹ non-coding region (3ʹ-UTR) of target gene messenger RNA (mRNA) so as to degrade it or repress its post-transcriptional translation.17,18 By regulating the expression of target genes, miRNA-186-5p, as a member of the microRNAs family, plays an important role in cell proliferation, apoptosis, differentiation and other processes, and in maintaining the normal function of cells.18,19 Therefore, miRNA disorders are associated with many diseases, such as tumors.19
Homeobox genes (HOX genes) are an important family of genes regulating embryonic development and cell growth and differentiation in vivo. With the continuous development of the understanding of HOX genes, recent studies have found that HOX genes are closely related to the occurrence, development, invasion and metastasis of tumors.20,21 Studies have shown that HOX gene plays a positive or negative regulatory role in the process of cell proliferation and differentiation, and is closely related to the occurrence and development of gastrointestinal tumors, breast cancer, endometrial cancer, etc.21 As a member of the HOX family, HOXA9 gene may be a new tumor molecular marker protein, which may play an important role in the development of ECa.22,23 Bioinformatics genome prediction suggested that HOXA9 may be one of the targeted genes of miRNA-186-5p. However, whether miRNA-186-5p can be involved in the proliferation and metastasis of ECa cells through the targeted regulation of HOXA9 is still unknown.
Therefore, this study further explored the biological function of microRNA-186-5p in ECa and the specific molecular mechanism of whether microRNA-186-5p could regulate HOXA9, providing clues to explain the potential mechanism of the occurrence and development of ECa, hoping to discover potential biomarkers for the early diagnosis and population screening of ECa.
Patients And Methods
Patients And ECa Samples
The tumor tissues and corresponding adjacent tissues were collected from forty-five patients who were diagnosed of ECa by pathology and underwent surgical resection. All samples were frozen and stored in a refrigerator at −80°C for subsequent RNA extraction. The study was approved by the Ethics Oversight Committee of Shandong Provincial Hospital Affiliated to Shandong University. All patients had not receive chemotherapy before surgery and the patients and their families were fully informed that the specimens would be used for scientific research and signed informed consent. This study was conducted in accordance with the Declaration of Helsinki. The pathological classification and staging criteria of ECa were performed according to the international association of cancer (UICC) ECa staging criteria.
Cell Lines And Reagents
Four human ECa cells (OE19, OE33, TE-1 and EC-109) and one human normal esophageal epithelial cell (HEEC) were purchased from ATCC, USA. Dulbecco’s modified eagle medium (DMEM, Thermo Fisher Scientific, Waltham, MA, USA) and fetal bovine serum (FBS) (Gibco, Rockville, MD, USA) were purchased from Life Technologies, USA. Cells were cultured at 37°C with 5% CO2 with MEM medium containing 10% FBS and 100 U/mL penicillin, 100 ug/mL streptomycin.
Cell Transfection
MicroRNA-186-5p overexpression was achieved by transfecting ECa cells TE-1 and EC-109 cell lines with microRNA-186-5p mimics (100 nmol/L). For recovery experiment, the HOXA9 overexpression plasmid was constructed and transfected into cells that had overexpressed microRNA-186-5p. Cells were divided into three groups: miR-NC+NC group, micro-186-5p mimics+NC group, microRNA-186-5p mimics+HOXA9 group. Cells were plated in 6-well plates and grown to a cell density of 70%, then plasmid transfection was performed using Lipofectamine 3000 (Invitrogen, Carlsbad, CA, USA) according to the manufacturer’s instructions, and cells were harvested 48 h later for qRT-PCR analysis and cell function experiments.
Cell Counting Kit (CCK-8) Assay
The cells after 48 h of transfection were harvested and plated into 96-well plates at 2000 cells per well. The cells were cultured for 24 h, 48 h,72 h and 96 h respectively, and then added with CCK-8 (Dojindo Laboratories, Kumamoto, Japan) reagent. After incubation for 2 hrs, the OD value of each well was measured in the microplate reader at 490 nm absorption wavelength.
Colony Formation Assay
After the cell density reached 70–80%, a single cell suspension was prepared by digesting with 0.25% trypsin. Subsequently, the cell suspension was repeatedly mixed using a pipette tip, and the number of viable cells was counted by the cell counting plate after trypan blue staining. 100 cells were seeded in a six-well plate, and 2 mL of complete medium was added and the plate was shaken for 5 min. Three parallel samples were set for each concentration, and the cells were cultured for 7 days in a 37 ° C, 5% CO2 incubator to calculate the colony formation rate.
Transwell Cell Migration Assay
After transfection for 48 hrs, the cells were trypsinized and resuspended in serum-free medium. After cell counting, the diluted cell density was adjusted to 2.0×105/mL, and the Transwell chamber containing Matrigel and no Matrigel was placed in a 24-well plate. 200 μL of the cell suspension was added to the upper chamber, and 500 μL of the medium containing 10% FBS was added to the lower chamber. After 48 hrs, the chamber was removed, fixed with 4% paraformaldehyde for 30 mins, and stained with the crystal violet for 15 mins. After washed with the PBS, the inner surface of the basement membrane of the chamber was carefully cleaned to remove the inner layer cells. The perforated cells stained in the outer layer of the basement membrane of the chamber were observed under the microscope, and 5 fields of view were randomly selected.
Quantitative Real-Time Polymerase Chain Reaction (qRT-PCR)
The transfected cells were cultured for 48 h, and RNA was extracted and reverse transcribed into cDNA using Primescript RT Reagent (TaKaRa, Otsu, Japan). QRT-PCR reactions were performed using SYBR® Premix Ex TaqTM (TaKaRa, Otsu, Japan), and StepOne Plus Real-time PCR System (Applied BiPCaystems, FP Cater City, CA, USA). The relative expression levels of microRNA-186-5p and HOXA9 were determined according to the methods described above. Data analysis was performed using ABI Step One software and the relative expression levels of mRNA were calculated using the 2−ΔΔCt method. All experiments were repeated 3 times.
Luciferase Report Gene Assay
According to the instructions, the esophageal cancer cell line in the logarithmic growth phase was prepared for the luciferase assay. A reporter plasmid was constructed in which a specific fragment of the target promoter was inserted in front of the luciferase expression sequence. The transcription factor expression plasmid to be detected was co-transfected with the reporter gene plasmid into the ECa cell line. If the transcription factor could activate the target promoter, the luciferase gene would be expressed, and the amount of luciferase expression was directly proportional to the intensity of the transcription factor. A specific luciferase substrate was added, and luciferase can react with the substrate to generate fluorescence. By measuring the intensity of the fluorescence, the activity of the luciferase can be calculated to determine whether the transcription factor can interact with the target promoter fragment.
Statistical Analysis
Data analysis was performed using Statistical Product and Service Solutions (SPSS) 22.0 (SPSS IBM, Armonk, NY, USA) statistical software. The measurement data were expressed as mean±standard deviation (x̅±s). The mean difference of microRNA-186-5p expression between groups was analyzed using paired sample test. The relationship of microRNA-186-5p and clinical pathology features was tested by χ2 test. The survival analysis was performed by Kaplan-Meier method, and the survival curve was drawn. P<0.05 was considered statistically significant.
Results
microRNA-186-5p Was Lowly Expressed In ECa Tissues And Cell Lines
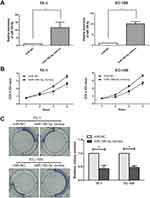
The expression of microRNA-186-5p in 45 pairs of ECa tissue specimens and adjacent ones as well as in cell lines was detected by qRT-PCR, and the results revealed that microRNA-186-5p was remarkably lower either in ECa tissues (P<0.001; Figure 1A) or cell lines (Figure 1B).
microRNA-186-5p Expression Was Correlated With Pathological Stage, Distant Metastasis And Overall Survival In ECa Patients
According to microRNA-186-5p expression, the 45 pairs of ECa tumor tissue samples and paracancerous ones were divided into high expression group and low expression group, and the relevant clinical indicators were analyzed. Chi-square test was used to analyze the relationship between microRNA-186-5p expression and age, gender, pathological stage, lymph node metastasis and distant metastasis of ECa patients. As shown in Table 1, the low expression of microRNA-186-5p was positively correlated with the incidence of clinical stage and distant metastasis of ECa, but not with age, gender and lymph node metastasis. In addition, in order to explore the interplay between microRNA-186-5p expression and prognosis in patients with ECa, we collected relevant follow-up data. Kaplan–Meier survival curves indicated that low expression of microRNA-186-5p was remarkably associated with poor prognosis of ECa, and the lower the expression level of microRNA-186-5p, the worse the prognosis (P<0.05; Figure 1C). These results suggested that microRNA-186-5p expression was correlated with pathological stage, distant metastasis and overall survival in ECa patients.
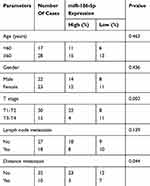 |
Table 1 Association of miR-186-5p expression with clinicopathologic characteristics of esophageal cancer |
Over-Expression Of microRNA-186-5p Inhibited Cell Proliferation
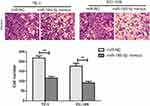
To explore the effect of microRNA-186-5p on the proliferation of ECa cells, microRNA-186-5p overexpression model was successfully constructed (P<0.01; Figure 2A). CCK-8 and plate cloning assays were performed to analyze proliferation ability of cells in the control miR-NC group and microRNA-186-5p mimics group. As shown in Figure 2B and C, the proliferative capacity of ECa cells in the microRNA-186-5p mimics group was remarkably reduced compared to the miR-NC group.
Over-Expression Of microRNA-186-5p Inhibited Cell Metastasis
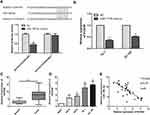
In order to explore the effect of microRNA-186-5p on the migration and metastasis of ECa cells, the migration and metastasis of the cells were detected by transwell migration assay in the miR-NC group and the microRNA-186-5p mimics group. The Transwell migration assay showed (P<0.01; Figure 3) that the microRNA-186-5p mimics group had a significant decrease in migration and metastasis of ECa cells compared with the miR-NC group.
microRNA-186-5p Could Bind To HOXA9
In order to further explore the way in which microRNA-186-5p regulated the malignant progression of ECa, we found that there may be a relationship between HOXA9 and microRNA-186-5p through relevant bioinformatics analysis. To further validate the targeting of microRNA-186-5p to HOXA9, a luciferase reporter assay was performed. The results showed that overexpression of microRNA-186-5p remarkably attenuated the luciferase activity of the wild-type HOXA9 vector, further demonstrating that HOXA9 can be targeted by microRNA-186-5p through this binding site (P<0.05; Figure 4A). Furthermore, by constructing the microRNA-186-5p overexpression vector, qRT-PCR detected the expression level of HOXA9 in the ECa cell line after overexpression of miR-188-5p, and the results indicated that the expression of HOXA9 was remarkably decreased (P<0.05; Figure 4B). Besides, qRT-PCR experiments showed that the expression level of HOXA9 was remarkably higher in ECa tissues, and the difference was statistically significant (P<0.001; Figure 4C). Meanwhile, HOXA9 was also remarkably higher in ECa cell lines than in HEEC, and the difference was statistically significant (Figure 4D). In addition, the expression of microRNA-186-5p and HOXA9 were detected by qRT-PCR, and the results showed that microRNA-186-5p and HOXA9 were negatively correlated in ECa tissues (Figure 4E). These results indicated that microRNA-186-5p could bind to HOXA9 and that there was a negative correlation between the expression of microRNA-186-5p and HOXA9.
Overexpression Of HOXA9 Reversed The Inhibitory Effect Of microRNA-186-5p In ECa Cells
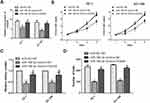
To further explore the interaction between microRNA-186-5p and HOXA9 in ECa cells, we overexpressed HOXA9 in a cell line overexpressing microRNA-186-5p in ECa cells to confirm there may be some mutual regulation between HOXA9 and HOXA9. Further, as shown in Figure 5A, the HOXA9 overexpression vector was constructed using a plasmid, and the transfection efficiency of HOXA9 after co-transfection of the microRNA-186-5p and HOXA9 vectors was examined by qRT-PCR. Subsequently, we demonstrated that overexpression of HOXA9 counteracted the effect of microRNA-186-5p mimics on proliferation and migration of ECa cells by cell CCK-8, plate cloning and Tranwell migration experiments (Figure 5B–D). These indicated that microRNA-186-5p modulated HOXA9 in ECa and that HOXA9 overexpression could reverse the inhibition effect of microRNA-186-5p on ECa cell proliferation and migration.
Discussion
Tumor refers to a new organism formed by cells in the body under the action of various tumorigenic factors, in which the cells in local tissues mutate at the gene level and lose the normal regulation of their growth, leading to unlimited abnormal proliferation and differentiation of cells.1–3 Once a tumor is formed, its growth characteristics are not regulated by normal body physiology, but destroy normal tissues and organs, especially in malignant tumors.4,5 Carcinoma refers to the malignant tumor that occurs in epithelial tissue and is caused by the abnormal growth of cells.6 In addition to their own unlimited proliferation, cancer cells will also be transferred to all parts of the body through the lymphatic system or circulatory system to grow and reproduce, resulting in cachexia and severe organ failure, etc., and eventually leading to the death of patients.6–8 Although great progress has been made in the diagnosis and treatment of ECa in the past decades, the prognosis of ECa, especially in patients with advanced and metastatic ECa, is still not satisfactory, and recurrence and metastasis are the main reasons for the failure of ECa treatment.8,9 Therefore, screening is an important method for early detection and early diagnosis of ECa, and improving the sensitivity and specificity of screening is the focus of current research.10–12
MicroRNA is a class of non-coding small molecule single-stranded RNA, which could regulate gene expression mainly at the transcriptional level and the post-transcriptional level of animals, plants, viruses and bacteria. By specifically binding to the base pair of target mRNA, microRNA can cause degradation or translation inhibition of mRNA and regulate gene expression.13,14 Many miRNAs have been found in animals, plants and viruses, and each miRNA regulates multiple target genes at the post-transcriptional level. To date, more than 700 human miRNAs have been cloned.15 Mature miRNA is a small mature RNA molecule with about 22 base pairs, which plays an important role in transcriptional regulation and is transformed from precursor miRNA(pre-microRNA).16–19 miRNA-186-5p is one of the newly discovered miRNAs in recent years, which can be expressed by targeting downstream genes and play a similar role of tumor suppressor genes.24,25 A large number of studies have confirmed that the low expression of miRNA-186-5p existed in a variety of malignant tumor tissues, and the low expression of microRNA-186-5p also provided favorable conditions for the proliferation and metastasis of malignant tumor cells to a certain extent.24–27 However, the effect of microRNA-186-5p on ECa is rarely reported. On the basis of previous studies, this chapter selected ECa cell lines TE-1 and EC-109 as the research subjects, and used cell transfection technology to explore the influence of miRNA-186-5p expression on the biological characteristics of TE-1 and EC-109 cell lines, such as proliferation, invasion and migration, and its regulatory effect on downstream target genes. qRT-PCR was used to verify the expression of miRNA-186-5p in ECa tumor tissues and adjacent tissues. The results showed that the expression of miRNA-186-5p was significantly decreased, and was positively correlated with the pathological staging, distant metastasis and poor prognosis of ECa. Therefore, we believed that miRNA-186-5p might play a role of tumor inhibition in ECa. In addition, through the construction of miRNA-186-5p transfection model, it was found that miRNA-186-5p mimics remarkably inhibited the proliferation, invasion and migration of the ECa cells TE-1 and EC-109, suggesting that miRNA-186-5p acted as a tumor suppressor gene in the occurrence and development of ECa.
The functional detection in the analysis of abnormal miRNA mainly focused on the mutations in target gene binding.16 These studies focused on the importance of complementary pairing between the 5 ‘end of miRNA and the 3ʹ UTRs region of target mRNA, and the complementary pairing region of miRNA and target mRNA is called the seed region.17,18 Computational studies after experimental verification emphasized the importance of seed region recognition in miRNA-target mRNA, and based on miRNA-target gene prediction and evolutionarily conserved sequences derived from experimental studies, it was estimated that more than 30% of animal genes may be the target sites of miRNA.19 At present, most studies have shown that HOXA9 was found in a variety of malignant tumors and might bind to the mRNA of miRNA-186-5p through bioinformatics. In this experiment, it was found through the recovery experiment that miRNA-186-5p and HOXA9 had mutual regulation effects. In order to further prove whether miRNA-186-5p promoted the development of ECa by regulating the HOXA9, we detected the down-regulated expression of HOXA9 after overexpression of miRNA-186-5p by qRT-PCR, revealing that miRNA-186-5p inhibited the proliferation and migration of ECa by regulating HOXA9. In addition, the results also showed that overexpression of HOXA9 could remarkably reverse the effect of miRNA-186-5p mimics in ECa, revealing that miRNA-186-5p might inhibit malignant progression of ECa by regulating HOXA9.
Conclusion
In summary, the expression of miRNA-186-5p in ECa was remarkably decreased, which was remarkably correlated with the pathological staging, distant metastasis and poor prognosis of ECa. Besides, miRNA-186-5p may inhibit the proliferation and metastasis of ECa by regulating HOXA9.
Disclosure
The authors report no conflicts of interest in this work.
References
1. de Gouw D, Klarenbeek BR, Driessen M, et al. Detecting pathological complete response in esophageal cancer after neoadjuvant therapy based on imaging techniques: a diagnostic systematic review and meta-analysis. J Thorac Oncol. 2019;14(7):1156–1171. doi:10.1016/j.jtho.2019.04.004
2. Verstegen M, Bouwense S, van Workum F, et al. Management of intrathoracic and cervical anastomotic leakage after esophagectomy for esophageal cancer: a systematic review. World J Emerg Surg. 2019;14:17. doi:doi:10.1186/s13017-019-0235-4
3. Zhang YG, Zhou MW, Bai L, Han RY, Lv K, Wang Z. Extracellular vesicles promote esophageal cancer progression by delivering lncZEB1-AS1 between cells. Eur Rev Med Pharmacol Sci. 2018;22(9):2662–2670. doi:doi:10.26355/eurrev_201805_14962
4. Amin G, Siegel M, Naimi T. National cancer societies and their public statements on alcohol consumption and cancer risk. Addiction. 2018;113(10):1802–1808. doi:10.1111/add.14254
5. Sardana RK, Chhikara N, Tanwar B, Panghal A. Dietary impact on esophageal cancer in humans: a review. Food Funct. 2018;9(4):1967–1977. doi:10.1039/c7fo01908d
6. Lian C, Xie S, Li W, et al. Association of wheat chaff derived silica fiber and esophageal cancer in north China. Ecotoxicol Environ Saf. 2019;178:79–85. doi:10.1016/j.ecoenv.2019.04.031
7. Sawada R, Maehara R, Oshikiri T, et al. MDM2 copy number increase: a poor prognostic, molecular event in esophageal squamous cell carcinoma. Hum Pathol. 2019;89:1–9. doi:10.1016/j.humpath.2019.04.002
8. Shen Y, Ding Y, Ma Q, et al. Identification of novel circulating miRNA biomarkers for the diagnosis of esophageal squamous cell carcinoma and squamous dysplasia. Cancer Epidemiol Biomarkers Prev. 2019;28(7):1212–1220. doi:10.1158/1055-9965.EPI-18-1199
9. Okamura A, Watanabe M, Kozuki R, et al. Significance of intramural metastasis in patients with esophageal squamous cell carcinoma: an indicator of aggressive cancer behavior. World J Surg. 2019;43(8):1997–2005. doi:10.1007/s00268-019-05004-z
10. Xia W, Liu S, Mao Q, et al. Effect of lymph node examined count on accurate staging and survival of resected esophageal cancer. Thorac Cancer. 2019;10(5):1149–1157. doi:10.1111/1759-7714.13056
11. Gao Z, Liu H, Shi Y, Yin L, Zhu Y, Liu R. Identification of cancer stem cell molecular markers and effects of hsa-miR-21-3p on stemness in esophageal squamous cell carcinoma. Cancers (Basel). 2019;11:4. doi:10.3390/cancers11040518
12. Hirano H, Kato K. Systemic treatment of advanced esophageal squamous cell carcinoma: chemotherapy, molecular-targeting therapy and immunotherapy. Jpn J Clin Oncol. 2019;49(5):412–420. doi:10.1093/jjco/hyz034
13. Su M, Xiao Y, Ma J, et al. Circular RNAs in cancer: emerging functions in hallmarks, stemness, resistance and roles as potential biomarkers. Mol Cancer. 2019;18(1):90. doi:10.1186/s12943-019-1002-6
14. Liu G, Li B. Role of miRNA in transformation from normal tissue to colorectal adenoma and cancer. J Cancer Res Ther. 2019;15(2):278–285. doi:10.4103/jcrt.JCRT_135_18
15. Mithraprabhu S, Morley R, Khong T, et al. Monitoring tumour burden and therapeutic response through analysis of circulating tumour DNA and extracellular RNA in multiple myeloma patients. Leukemia. 2019;33:2022–2033. doi:10.1038/s41375-019-0469-x
16. Dharmawardana N, Ooi EH, Woods C, Hussey D. Circulating microRNAs in head and neck cancer: a scoping review of methods. Clin Exp Metastasis. 2019;36(3):291–302. doi:10.1007/s10585-019-09961-6
17. Takahashi RU, Prieto-Vila M, Kohama I, Ochiya T. Development of miRNA-based therapeutic approaches for cancer patients. Cancer Sci. 2019;110(4):1140–1147. doi:10.1111/cas.13965
18. Tassinari V, Cesarini V, Silvestris DA, Gallo A. The adaptive potential of RNA editing-mediated miRNA-retargeting in cancer. Biochim Biophys Acta Gene Regul Mech. 2019;1862(3):291–300. doi:10.1016/j.bbagrm.2018.12.007
19. Tesfaye AA, Azmi AS, Philip PA. miRNA and gene expression in pancreatic ductal adenocarcinoma. Am J Pathol. 2019;189(1):58–70. doi:10.1016/j.ajpath.2018.10.005
20. Qu X, Alsager S, Zhuo Y, Shan B. HOX transcript antisense RNA (HOTAIR) in cancer. Cancer Lett. 2019;454:90–97. doi:10.1016/j.canlet.2019.04.016
21. Primon M, Hunter KD, Pandha HS, Morgan R. Kinase regulation of HOX transcription factors. Cancers (Basel). 2019;11:4. doi:10.3390/cancers11040508
22. Yu SL, Koo H, Lee HY, Yeom YI, Lee DC, Kang J. Recombinant cell-permeable HOXA9 protein inhibits NSCLC cell migration and invasion. Cell Oncol (Dordr). 2019;42(3):275–285. doi:10.1007/s13402-019-00424-4
23. Lynch JR, Salik B, Connerty P, et al. JMJD1C-mediated metabolic dysregulation contributes to HOXA9-dependent leukemogenesis. Leukemia. 2019;33(6):1400–1410. doi:10.1038/s41375-018-0354-z
24. Feng H, Zhang Z, Qing X, French SW, Liu D. miR-186-5p promotes cell growth, migration and invasion of lung adenocarcinoma by targeting PTEN. Exp Mol Pathol. 2019;108:105–113. doi:10.1016/j.yexmp.2019.04.007
25. Zhang Z, Zhang W, Mao J, Xu Z, Fan M. miR-186-5p functions as a tumor suppressor in human osteosarcoma by targeting FOXK1. Cell Physiol Biochem. 2019;52(3):553–564. doi:10.33594/000000039
26. Li J, Xia L, Zhou Z, et al. MiR-186-5p upregulation inhibits proliferation, metastasis and epithelial-to-mesenchymal transition of colorectal cancer cell by targeting ZEB1. Arch Biochem Biophys. 2018;640:53–60. doi:10.1016/j.abb.2018.01.002
27. Islam F, Gopalan V, Vider J, et al. MicroRNA-186-5p overexpression modulates colon cancer growth by repressing the expression of the FAM134B tumour inhibitor. Exp Cell Res. 2017;357(2):260–270. doi:10.1016/j.yexcr.2017.05.021
 © 2019 The Author(s). This work is published and licensed by Dove Medical Press Limited. The full terms of this license are available at https://www.dovepress.com/terms.php and incorporate the Creative Commons Attribution - Non Commercial (unported, v3.0) License.
By accessing the work you hereby accept the Terms. Non-commercial uses of the work are permitted without any further permission from Dove Medical Press Limited, provided the work is properly attributed. For permission for commercial use of this work, please see paragraphs 4.2 and 5 of our Terms.
© 2019 The Author(s). This work is published and licensed by Dove Medical Press Limited. The full terms of this license are available at https://www.dovepress.com/terms.php and incorporate the Creative Commons Attribution - Non Commercial (unported, v3.0) License.
By accessing the work you hereby accept the Terms. Non-commercial uses of the work are permitted without any further permission from Dove Medical Press Limited, provided the work is properly attributed. For permission for commercial use of this work, please see paragraphs 4.2 and 5 of our Terms.