Back to Journals » Cancer Management and Research » Volume 13
MicroRNA-148a-3p Directly Targets SERPINE1 to Suppress EMT-Mediated Colon Adenocarcinoma Progression
Authors Hu B, Chen Z, Wang X , Chen F , Song Z , Cao C
Received 20 January 2021
Accepted for publication 5 June 2021
Published 11 August 2021 Volume 2021:13 Pages 6349—6362
DOI https://doi.org/10.2147/CMAR.S302777
Checked for plagiarism Yes
Review by Single anonymous peer review
Peer reviewer comments 2
Editor who approved publication: Dr Eileen O'Reilly
Biwen Hu,* Zhenwei Chen,* Xiaoguang Wang, Fei Chen, Zhengwei Song, Chenxi Cao
Department of Surgery, The Second Affiliated Hospital of Jiaxing University, Jiaxing, Zhejiang Province, People’s Republic of China
*These authors contributed equally to this work
Correspondence: Chenxi Cao
Department of Surgery, The Second Affiliated Hospital of Jiaxing University, No. 397, Huangcheng North Road, Jiaxing, Zhejiang, 314000, People’s Repubic of China
Tel +86 573 82058116
Email [email protected]
Aim: This research aimed at clarifying the intracellular effect of SERPINE1 in the progression of colon adenocarcinoma (COAD) and the underlying mechanism.
Methods: We obtained the expression profile of SERPINE1 in COAD via the Starbase database and verified it on COAD tissue samples through qRT-PCR and immunoblotting, respectively. Also, miRWalk, TargetScan and miRDB databases were adopted to generate the miRNA prediction that might target SERPINE1, and the gene target miR-148a-3p was confirmed using dual-luciferase assays. The effect of SERPINE1 and miR-148a-3p on COAD was further evaluated by cell experiments. MTT assay was used to detect the change of cell proliferation ability. The invasive and migratory capability of COAD cells was examined using transwell and would healing assays. Cell apoptosis was determined through flow cytometry. The expressions of genes and EMT-associated proteins were evaluated by qRT-PCR and immunoblotting. Further lucubration of the biological relevance of SERPINE1 and miR-148a-3p was conducted using rescue experiments.
Results: We found that the expression quantities of SERPINE1 in COAD tissues and cell lines were higher than those in corresponding non-cancerous tissues and normal cells. When SERPINE1 expression is reduced, EMT process is inhibited, invasion and proliferation ability of COAD cells are obviously reduced, and apoptosis level is increased. Moreover, SERPINE1 was identified as the target gene of miR-148a-3p. When the expression of miR-148a-3p was enhanced, it was found that the expression of SERPINE1 was reduced. miR-148a-3p played the similar effect of si-SERPINE1 that suppressed the COAD progression. Additionally, we found out that SERPINE1 is validated in hindering the tumor healing effect of miR148a-3p in COAD, including cell growth and invasion.
Conclusion: Our study suggests that SERPINE1/miR-148a-3p axis has potential as prognostic markers of COAD and provides reference for the development of new therapies.
Keywords: colon adenocarcinoma, SERPINE1, miR-148a-3p, EMT
Introduction
Cancer is a common cause of death worldwide (8.97 million deaths) after ischemic heart disease.1 Colon adenocarcinoma (COAD) is a common malignant cancer, resulting in high mortality2 with a proportion of 5%.3 Currently, although many treatment approaches have increased the life quality of patients with COAD, the 5-year survival rate is still poor,4,5 Late diagnosis and lack of effective therapeutic targets are the main obstacles to COAD treatment.6,7 Hence, reliable biomarkers and specific therapeutic targets affirmatively help improve diagnosis and decrease death rate in COAD patients.8,9 Therefore, it is urgent to investigating the molecular mechanism of COAD tumorigenesis, which may provide new treatment targets and prognostic biomarkers.
SERPINE1 belongs to the serine protein kinase inhibitor family, and its encoded protein that precluding the fibrinolysis process.10 Recent study reported that SERPINE1 was overexpressed in colorectal carcinoma as well as head and neck cancer, which indicated the pathological correlation of SERPINE1 and colorectal tumorigenesis.11 Also, SERPINE1 has been reported as a pro-tumorigenesis factor and a member of EMT pathway, which suggested that SERPINE1 enabling tumor cell growth and invasion.12,13 Some studies have also declared that the increase of SERPINE1 level in tumor and serum of COAD patients has a pathological association with poor prognosis, which can possibly be used as tumor or serum marker to more accurately determine the stage of COAD patients.14 Therefore, further exploration of the specific role of SERPINE1 in COAD would be of great help for improving COAD therapeutic approaches.
MicroRNAs (miRNAs) are currently considered as a key factor in cell differentiation, cell cycle and cell apoptosis by regulating mRNA, which is an important regulating factor of tumorigenesis.15 Many researchers have reported that aiming at miRNAs may be an alternative manner to block tumor progression. MiR-148a-3p participates in regulating various procedures in bladder carcinoma through preventing EMT, such as inhibiting migration and invasion.16 miR-148a-3p was predicted as the target miRNA of SERPINE1 by databases. Additionally, miR-148-3p is recognized as a tumor-suppressive miRNA which has an ability to preclude cell proliferation in laryngeal squamous cell carcinoma (LSCC).17 Hence, we hypothesized that miR-148a-3p might have a function inhibiting COAD, which could help further understand the mechanism of tumor progression, including cell growth and invasion in COAD.
Here, we lucubrated the specific function of intracellular SERPINE1 on COAD and the detailed mechanisms of its pro-tumorigenic ability. We discovered that SERPINE1 was negatively associated with the prognosis of COAD patients. After SERPINE1 was silenced, EMT process was obviously inhibited, invasion ability of COAD cells decreased, apoptosis was promoted, and cell proliferation was inhibited. MiR-148-3p is positively correlated with the prognosis of COAD patients, and SERPINE1 is targeted to exert its anti-tumor effect. Therefore, we have proposed a theory that miR-148-3p targets SERPINE1, thus inhibiting EMT process and COAD progression.
Materials and Methods
Bioinformatics Analysis
TCGA is a central repository of multidimensional experimental cancer data, comprising data pertaining to >30 types of human tumors. We obtained the expression profile of SERPINE1 in COAD and corresponding non-cancerous tissues via the Gene Expression Profiling Interactive Analysis (Starbase) tool (http://starbase.sysu.edu.cn/index.php) based on TCGA database.
We used miRWalk, TargetScan and miRDB databases to predict miRNAs that may target SERPINE1.
Patients and Tissue Samples
A total of 10 pairs of colorectal carcinoma tissues and adjacent normal tissues from patients with COAD who had undergone resection between 2004 and 2013 in the Second Affiliated Hospital of Jiaxing University. All samples were immediately frozen and ground using liquid nitrogen, and subsequently stored under −80°C till RNA extraction. The study was approved by the Ethics Committee of Jiaxing University and conducted in accordance with the Declaration of Helsinki, and all subjects signed informed consent.
Cell Culture
Human COAD cell lines (SW480, RKO, HT29 and HTC116) (American Type Culture Collection, Manassas, VA, USA) and NCM460 human colon epithelial (the Second Affiliated Hospital of Jiaxing University) were cultured in DMEM medium (Gibco, USA) containing 10% fetal bovine serum (FBS; Gibco) at 37°C and 5% CO2. The NCM460 cell lines was approved by the Ethics Committee of Jiaxing University.
Total RNA Extraction and qRT-PCR
Human colorectal cancer tissues and colorectal cell lines were collected to extract the RNA for continual experiment. In order to lyse cells and avoid RNA lysis, 1 mL Trizol reagent (Invitrogen, Carlsbad, CA, USA) was added to cell and tissue suspension. Reverse transcription reagents (Thermo Scientific, Waltham, MA, USA) were utilized to completed cDNA synthesis. Additional chloroform at room temperature was added to the mixture, then added isopropanol to centrifugate to get RNA precipitate. Real-time qPCR of SERPINE1, miR-148-3p and EMT-related transcripts was conducted through an ABI ViiA 7 sequence detection system (ABI, Carlsbad, CA, USA) and a SYBR Premix Ex Taq™ II kit (TaKaRa Bio, Inc., Shiga, Japan) based on the manufacturer's instruction. The transcription level was calculated by 2−ΔΔCt method.18
Small Interfering RNA Transfection and pcDNA3.1 Transfection
Small interfering RNA targeting SERPINE1 (si-SERPINE1), the control sequences (si-NC), miR-148a-3p mimics and its corresponding negative control (NC) were synthesized by Genepharma (Shanghai, China) and were then amplified and transfected into COAD cells to construct a SERPINE1 knockdown cell line. Simultaneously, the negative control shRNA was also co-incubated with SW480 or HCT116. Conversely, pcDNA3.1-SERPINE1 was applied for the up-regulation of SERPINE1, of which blank pcDNA3.1 was utilized as related negative control. Additionally, miR-148a-3p upregulated cell line was constructed by transfecting miR-148a-3p mimics to SW480 for the continual experiments.
Western Blotting
Cells were collected and lysed in lysis buffer on ice for 30 min. Subsequently, the protein concentration was determined using ultraviolet spectrophotometer (Thermo Scientific, Waltham, MA, USA). Proteins solution was then added loading buffer and boiled for 5 min to be fully denatured and transferred onto polyvinyldifluoride membranes (Bio-Rad Laboratories USA) after separated on SDS-PAGE. The membranes were immersed with the diligent of SERPINE1, miR-148a-3p and EMT-related proteins’ antibodies, and then observed using an enhanced chemiluminescence reagent (Thermo Scientific, Waltham, MA, USA).
Dual-Luciferase Assay
The wild-type SERPINE1-1 3ʹ-UTR (WT) was amplified by RT-PCR and cloned into the psiCHECK-2 vector. Site-Directed Mutagenesis kit (Invitrogen, Carlsbad, CA, USA) was adopted to generate SERPINE1 site mutation, and then constructed mutated SERPINE1 (Mut) reporter plasmid in the same way. The SERPINE1-related plasmid and the control plasmid were then co-transfected with miR-148a-3p mimics or inhibitor into SW480 cells. Then dual-Luciferase Reporter Assay System (Promega Corporation, Madison, WI, USA) was utilized to examine the luciferase activity.
Transwell Invasion Assay
The 24-well Transwell chamber (8 μm; Corning Incorporated, Corning, NY, USA) was used for the invasion assays. Cells transfected for 48h were collected, and the cell concentration was adjusted to 105 cells/mL. One hundred microliters of cell suspension was inoculated in transwell chamber. Twenty-four hours after seeding, the invasive ability of cancer cells was determined by the number of invasive cells that stained with crystal violet and photographed under a microscope.
Cell Viability Examination Through MTT Assay
Cells (SW480) were planted in 96-well plate for 5000 cells/100 μL per well. After 48 h of transfection, the excess medium was sucked out, then 80 μL of fresh medium was added, 20 μL of MTT was added and cultured for 4 h. Finally, the OD values were measured at A495 nm and A630 nm by enzyme-linked universal microplate spectrophotometer.
Flow Cytometry for Apoptosis Assay
The cells were incubated with si-SERPINE1 and miR-148a-3p mimics for 6 hours. Then, the transfected cells grew in fresh medium for 72 h. 5×105 transfected cells were collected and subsequently suspended in 100 μL buffer. Add 5μL annexin V and 5μL 7-AAD and incubate in dark at 25°C for 15min. Finally, add 400 μL of combined buffer, place it on ice (keep away from light), and were tested using flow cytometry within 1h.
Wound Healing Assay
Wound healing assay is a method to detect cell migration in vitro, which is widely used in the measurement of cell migration ability in vitro. HCT116 and SW480 cells in logarithmic growth phase were collected, and the cell concentration was adjusted to 3×105 cells/mL, and inoculated into 6-well plate for 24 h of incubation in the incubator. After the cells formed monolayer fusion, the cells were scratched with sterile 10 μL of pipette tip perpendicular to the horizontal line in each hole. Mark the horizontal line evenly on the back of 6-hole plate with marking pen and ruler. The width of the scratch was recorded. ImageJ software was used to measure the scratch area at different times, and GraphPad software was used to draw and analyze the results.
Statistical Studies
The results’ data were expressed as mean of triplicate measurements plus standard deviation (SD). Statistically significant differences in different groups were evaluated by Student’s t test through SPSS (13.0) or GraphPad Prism 7.0. P < 0.05 means statistically significance.
Ethics Approval and Consent to Participate
All clinical procedures are in accordance with current clinical guidelines and regulations. This study was permitted by the Ethics Committee of the Second Affiliated Hospital of Jiaxing University.
Results
SERPINE1 is Highly Expressed in COAD
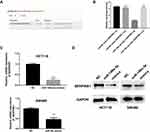
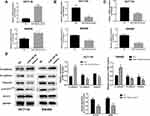
The results of Starbase database showed that SERPINE1 was highly expressed in COAD (Figure 1A). Real-time qPCR and Western blot were then conducted to verify the expression of SERPINE1 in COAD tissue samples from 10 patients. Consistent with the consequence in Starbase database, the expression level of SERPINE1 in 10 COAD tissue was higher than that in the corresponding non-cancerous tissues (Figure 1B and C).
Knock Down of SERPINE1 Repressed EMT-Mediated Metastasis of COAD Cells
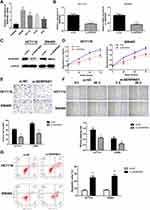
SERPINE1 expression level measured using qRT-PCR in human colorectal cancer cell lines (SW480, RKO, HT29 and HTC116) were significantly higher than that in NCM460 cells (Figure 2A), and SW480 and HCT116 were discovered as the highest SERPINE1 expressed cell line and was utilized in the following experiments. To study the specific function of SERPINE1 on COAD, we established SW480 and HCT116 cells stably knocking down SERPINE1 through lentiviral infection and the transfection efficiency was measured using qRT-PCR and Western blot assay (Figure 2B and C). Additionally, MTT assay was exerted to estimated cell viability of SERPINE1 knocking down cells. Results showed that knockdown of SERPINE1 could significantly decrease cell growth of HCT116 and SW480 (Figure 2D).
Next, the migratory and invasive capability was detected through transwell assay and wound healing assay. Compared with control cells, suppressive cell invasive capacity was exhibited in cells of si-SERPINE1 group (Figure 2E). Similarly, decreased wound healing ratio in si-SERPINE1 transfected HCT116 and SW480 cells suggested the alleviated cell migration in COAD (Figure 2F). Previous studies have reported that EMT has anti-apoptosis effect.19,20 Therefore, we also detected the apoptosis of the above cells with flow cytometry. We found that after SERPINE1 knocking down, apoptosis of SW480 and HCT116 cells synchronously increased. The experimental consequence demonstrated that knocking down SERPINE1 led to higher cell apoptosis rate both in HCT116 and SW480 cells (Figure 2G).
Epithelial-mesenchymal transition (EMT) is pathologically associated with the invasion and metastasis of cancer cells,21,22 and is recognized as a crucial factor for cell metastasis.23 The expressions of E-cadherin, N-cadherin and vimentin proteins were estimated to examine whether SERPINE1 affected EMT in COAD cells.24 As depicted in Figure 3A–D, knockdown of SERPINE1 decreased N-cadherin and 'vimentin and increased E-cadherin expression in SW480 and HCT116 cells detected by qRT-PCR and immunoblotting (Figure 3A–D), which indicated that SERPINE1 has the ability to enhance COAD cell invasion via promoting the EMT procedure. Thus, we considered that knockdown of SERPINE1 inhibits EMT-mediated metastasis of COAD cells.
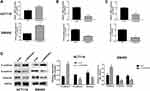
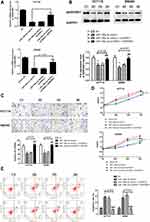
miR-148a-3p Was Confirmed as a Directly Target of SERPINE1
miRWalk, TargetScan and miRDB databases were adopted to give an intersection prediction of the miRNAs, which indicated that miR-148a-3p might directly target SERPINE1. The binding site of SERPINE1 and miR-148a-3p was shown in Figure 4A. Luciferase reporter assays further verified that SERPINE1 is a direct target of miR-148a-3p (Figure 4B). Subsequently, the concrete relevance between miR-148a-3p and SERPINE1 was determined through qRT-PCR and immunoblotting assay, and the results demonstrated that overexpression of miR-148a-3p using miR-148a-3p mimics induced lower-than-usual mRNA and proteins quantities of SERPINE1 both in HCT116 and SW480 cells (Figure 4C and D). These consequences indicated that SERPINE1 is negatively regulated by miR-148a-3p, thereby mediating the developed progress of COAD.
miR-148a-3p Directly Targets SERPINE1 to Inhibit EMT-Mediated COAD Cell Metastasis, and Hindered Cell Proliferation by Potentiating Apoptosis via Silencing STAT3
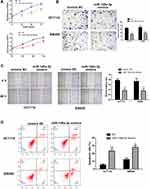
In order to examine the specific role of miR-148a-3p in COAD cell metastasis, we constructed stable miR-148a-3p overexpressing cells by transfecting miR-148a-3p mimics into SW480 and HCT116 cells. The cell viability using MTT assay exhibited that COAD cells (HCT116 and SW480) with high quantity of miR-148a-3p resulted in significant reduction of cell viability when compared with mimics NC group (Figure 5A), which indirectly elucidated the inhibitory function of miR-148a-3p in cell growth of COAD.
Next, we examined the function of miR-148a-3p on the invasive ability of COAD cells (HCT116 and SW480) using transwell assay. The results showed that cell invasion was reduced both in SW480 and HCT116 cells with miR-148a-3p overexpression (Figure 5B). Moreover, the research consequence of wound healing assay showed that overexpression of miR-148a-3p led to conspicuous alleviation of wound healing ratio both in HCT116 and SW480 cells (Figure 5C), which indirectly illustrated miR-148a-3p weaken cell migration ability of COAD. Besides, cell apoptosis alteration with miR-148a-3p overexpression was detected by flow cytometry. The consequences demonstrated that overexpression of miR-148a-3p promoted COAD cell apoptosis (Figure 5D).
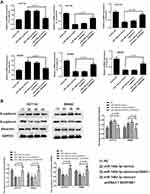
Consistently, up-regulation of miR-148a-3p induced increased E-cadherin, and decreased E-cadherin and vimentin expression in SW480 and HCT116 cells (Figure 6A–D). Moreover, we selected one of the typical tumor regulatory proteins STAT3 that closely participates in the tumorigenic process. Our finding showed that overexpression of miR-148a-3p resulted in down-regulation of phosphorylation of STAT3 with no alteration of total STAT3, which indicated that miR-148a-3p was capable to suppress the activation of STAT3 and thereby hindered the progress of COAD.
Consequently, we speculated that after miR-148a-3p targeted inhibition of SERPINE1, EMT process is inhibited, resulting in simultaneous increase of apoptosis and inhibition of cell proliferation. Also, our findings show that miR-148a-3p inhibits EMT-mediated metastasis in COAD by directly targeting SERPINE1.
SERPINE1 Reversed the Effect of miR-148a-3p on the Development of COAD Cells
Exploration of the concrete modulatory relation of miR-148a-3p and SERPINE1 in COAD was continued by examining the rescue experiments. MiR-148a-3p was given to both HCT116 and SW480 cells to establish the overexpression of miR-148a-3p and down-regulation of SERPINE1, since we have determined that SERPINE1 was down-regulated when miR-148a-3p was overexpressed. Also, pcDNA3.1-SERPINE1 was co-treated with miR-148a-3p to further up-regulate the quantity of SERPINE1 in COAD. Depicted in Figure 7A was the transfection efficiency of 4 groups, which elucidated the successful cell transfection both in HCT116 and SW480 cells. Immunoblotting assay was also performed to determine the SERPINE1 expression level and found that miR-148a-3p mimics treated group led to SERPINE1 reduction, while further transfection of pcDNA3.1-SERPINE1 converses the result to high quantity of SERPINE1 (Figure 7B).Subsequently, we estimated cell invasion of COAD of these four groups by transwell assay. Consistent with our speculation, miR-148a-3p mimics treated group presented lower cell invasion while up-regulation of SERPINE1 reversed the effect of miR-148a-3p (Figure 7C). Cell viability was also determined using MTT assay in HCT116 and SW480 cells, and our results showed that solo miR-148a-3p mimics led to obvious lower cell viability versus co-treatment of miR-148a-3p mimics and pcDNA3.1-SERPINE1 group (Figure 7D). Further, related biological mechanism of this effect was examined using FCM, and we found that overexpression of miR-148a-3p induced higher apoptosis rate versus negative control. Yet overexpression of SERPINE1 could reverse the effect of miR-148a-3p and suppressed cell apoptosis (Figure 7E). Next, further study the mechanism of cell invasion was conducted using qRT-PCR and Western blot. The results of these two assays altogether illustrated that miR-148a-3p mimics led to up-regulation of E-cadherin and down-regulation of N-cadherin as well as vimentin both at mRNA and protein levels in HCT116 and SW480 cells, while overexpression of SERPINE1 presented the opposite expression situation (Figure 8A and B).
Collectively, these consequences illustrated that SERPINE1 would reverse the tumor suppressive effect of miR-148a-3p, involving cell growth and invasion.
Discussion
Adenocarcinoma is the main precancerous disease of colorectal cancer, accounting for 85% - 90% of all precancerous diseases in colorectal cancer. Most colorectal cancer was in middle and late stage when diagnosed, so early diagnosis is very important for the treatment of colorectal cancer.25
SERPINE1, also known as Plasminogen activator inhibitor 1 (PAI-1), is recently recognized as a promoting factor of various cancers, including esophageal cancer, prostate cancer and gastric cancer.26–28 Current researches identified SERPINE1 as a participant in tumor cell progression, involving proliferation, metastasis and invasion. Previous studies have shown that SERPINE1 is associated with poor prognosis of COAD patients,14 which may promote tumor growth through EMT pathway.29 However, the mechanism of intracellular SERPINE1 in COAD progression has not been determined. Therefore, identifying the specific mechanism of SERPINE1 in COAD might help discover novel approaches to improve COAD therapy and prognosis.
To lucubrate the SERPINE1 expression level in COAD, we utilized TCGA database for expression analyses and found that SERPINE1 is highly expressed in COAD. Then, further detecting of the expression level of SERPINE1 in COAD clinical tissues were performed through qRT-PCR and Western blotting, confirming that SERPINE1 expression in COAD tissues is higher than that in corresponding non-cancerous tissues.
In order to verify the role and mechanism of SERPINE1 in COAD progression, we further carried out Real-time qPCR, Western blot, transwell invasion assay, MTT assay, flow cytometry and other experiments. Investigation certified that SERPINE1 could promote cell proliferation through preventing apoptosis of COAD. Also, evidences prove that SERPINE1 plays an active role in the accelerating cell invasion of COAD via activating EMT by examining EMT-related proteins through Western blot. Knockdown of SERPINE1 gene inhibited EMT process, reduces cell invasion and proliferation ability and promotes cell apoptosis.
Recently, miR-148a-3p is found as a modulating factor in various cancer including gastric cancer,30 laryngeal squamous cell carcinoma31 and colon tumor.32 The exact function of miR-148a-3p in regulating tumor progression has some nuances in different oncology. For example, by direct targeting c-Met, miR-148a-3p preclude metastasis of malignant epithelial ovarian carcinoma (EOC).33 When miR-148a-3p serves as a tumor suppressor in EOC, it obviously showed similar function in osteosarcoma,34 overexpression of miR-148a-3p significantly reduced cell growth and promoted apoptosis. To further illustrate the function of miR-148a-3p in COAD, we next study whether miR-148a-3p also presents similar tumor repressing effect in promoting apoptosis and preventing cell invasion. Here, SERPINE1 has been discovered as the direct target of miR-148a-3p through dual-luciferase assay, which suggested that miR-148a-3p is also a specific factor modulating cell progression of COAD. Further exploration of miR-148a-3p in the process of cell growth and invasion illustrated that miR-148a-3p can directly target SERPINE1 and exert its anti-cancer effect by inhibiting EMT process. The rescue experiments also demonstrated this consequence (Figs.7 and 8). Additionally, in this study, we discovered that miR-148a-3p also partakes in suppressing the activation of STAT3 from Western blot assay. Consequently, we draw a conclusion that miR-148a-3p/SERPINE1 axis is a novel regulatory factor in COAD, which provides new strategy for COAD therapeutic methods and early diagnosis.
Conclusion
In conclusion, our research shows that miR-148a-3p inhibits EMT-mediated COAD progression by directly targeting SERPINE1. SERPINE1 and miR-148a-3p have the potential to be prognostic markers of COAD and provide reference for the development of new therapies, but further experiments are needed to confirm this point.
Acknowledgments
We sincerely thank the participants who included in our research, and the science and technology planning project of Jiaxing City (No. 2018AY32002) and the science and technology planning project of Jiaxing City (No. 2019AD32254).
Disclosure
The authors declare no conflicts of interest in this study.
References
1. Mattiuzzi C, Lippi G. Current cancer epidemiology. J Epidemiol Glob Health. 2019;9(4):217–222. doi:10.2991/jegh.k.191008.001
2. Phang CW, Karsani SA, Abd Malek SN. Induction of apoptosis and cell cycle arrest by Flavokawain C on HT-29 human colon adenocarcinoma via enhancement of reactive oxygen species generation, upregulation of p21, p27, and GADD153, and inactivation of inhibitor of apoptosis proteins. Pharmacogn Mag. 2017;13(Suppl 2):S321–s328.
3. Bray F, Ferlay J, Soerjomataram I, Siegel RL, Torre LA, Jemal A. Global cancer statistics 2018: GLOBOCAN estimates of incidence and mortality worldwide for 36 cancers in 185 countries. CA Cancer J Clin. 2018;68(6):394–424.
4. Hong J, Lu H, Meng X, Ryu JH, Hara Y, Yang CS. Stability, cellular uptake, biotransformation, and efflux of tea polyphenol (-)-epigallocatechin-3-gallate in HT-29 human colon adenocarcinoma cells. Cancer Res. 2002;62(24):7241–7246.
5. Roncucci L, Mariani F. Prevention of colorectal cancer: how many tools do we have in our basket? Eur J Intern Med. 2015;26(10):752–756.
6. Azar I, Esfandiarifard S, Sinai P, Mehdi S. Colon cancer survival in the US Department of Veterans Affairs by race and stage: 2001 through 2009. Cancer. 2018;124(13):2858.
7. Tsukuda K, Tanino M, Soga H, Shimizu N, Shimizu K. A novel activating mutation of the K-ras gene in human primary colon adenocarcinoma. Biochem Biophys Res Commun. 2000;278(3):653–658.
8. Tsimberidou AM. Targeted therapy in cancer. Cancer Chemother Pharmacol. 2015;76(6):1113–1132.
9. Herzig DO, Tsikitis VL. Molecular markers for colon diagnosis, prognosis and targeted therapy. J Surg Oncol. 2015;111(1):96–102.
10. Binder BR, Christ G, Gruber F, et al. Plasminogen activator inhibitor 1: physiological and pathophysiological roles. News Physiol Sci. 2002;17:56–61.
11. Mazzoccoli G, Pazienza V, Panza A, et al. ARNTL2 and SERPINE1: potential biomarkers for tumor aggressiveness in colorectal cancer. J Cancer Res Clin Oncol. 2012;138(3):501–511.
12. Placencio VR, DeClerck YA. Plasminogen Activator Inhibitor-1 in Cancer: rationale and Insight for Future Therapeutic Testing. Cancer Res. 2015;75(15):2969–2974.
13. Rokavec M, Oner MG, Li H, et al. IL-6R/STAT3/miR-34a feedback loop promotes EMT-mediated colorectal cancer invasion and metastasis. J Clin Invest. 2014;124(4):1853–1867.
14. Berger DH. Plasmin/plasminogen system in colorectal cancer. World J Surg. 2002;26(7):767–771.
15. Lujambio A, Lowe SW. The microcosmos of cancer. Nature. 2012;482(7385):347–355.
16. Wang X, Liang Z, Xu X, et al. miR-148a-3p represses proliferation and EMT by establishing regulatory circuits between ERBB3/AKT2/c-myc and DNMT1 in bladder cancer. Cell Death Dis. 2016;7(12):e2503.
17. Wu T, Qu L, He G, et al. Regulation of laryngeal squamous cell cancer progression by the lncRNA H19/miR-148a-3p/DNMT1 axis. Oncotarget. 2016;7(10):11553–11566.
18. Ginzinger DG. Gene quantification using real-time quantitative PCR: an emerging technology hits the mainstream. Exp Hematol. 2002;30(6):503–512.
19. Garg M. Epithelial plasticity, autophagy and metastasis: potential modifiers of the crosstalk to overcome therapeutic resistance. Stem Cell Rev Rep. 2020.
20. Na TY, Schecterson L, Mendonsa AM, Gumbiner BM. The functional activity of E-cadherin controls tumor cell metastasis at multiple steps. Proc Natl Acad Sci U S A. 2020;117(11):5931–5937.
21. Thiery JP, Acloque H, Huang RY, Nieto MA. Epithelial-mesenchymal transitions in development and disease. Cell. 2009;139(5):871–890.
22. Nauseef JT, Henry MD. Epithelial-to-mesenchymal transition in prostate cancer: paradigm or puzzle? Nat Rev Urol. 2011;8(8):428–439.
23. Kim MS, Lee WS, Jeong J, Kim SJ, Jin W. Induction of metastatic potential by TrkB via activation of IL6/JAK2/STAT3 and PI3K/AKT signaling in breast cancer. Oncotarget. 2015;6(37):40158–40171.
24. Lamouille S, Xu J, Derynck R. Molecular mechanisms of epithelial-mesenchymal transition. Nat Rev Mol Cell Biol. 2014;15(3):178–196.
25. Binefa G, Rodriguez-Moranta F, Teule A, Medina-Hayas M. Colorectal cancer: from prevention to personalized medicine. World J Gastroenterol. 2014;20(22):6786–6808.
26. Klimczak-Bitner AA, Kordek R, Bitner J, Musial J, Szemraj J. Expression of MMP9, SERPINE1 and miR-134 as prognostic factors in esophageal cancer. Oncol Lett. 2016;12(5):4133–4138.
27. Guo K, Zheng S, Xu Y, Xu A, Chen B, Wen Y. Loss of miR-26a-5p promotes proliferation, migration, and invasion in prostate cancer through negatively regulating SERBP1. Tumour Biol. 2016;37(9):12843–12854.
28. Yang JD, Ma L, Zhu Z. SERPINE1 as a cancer-promoting gene in gastric adenocarcinoma: facilitates tumour cell proliferation, migration, and invasion by regulating EMT. J Chemother. 2019;31(7–8):408–418.
29. Tomita T, Ieguchi K, Coin F, et al. ZFC3H1, a zinc finger protein, modulates IL-8 transcription by binding with celastramycin A, a potential immune suppressor. PLoS One. 2014;9(9):e108957.
30. Song B, Du J, Song DF, Ren JC, Feng Y. Dysregulation of NCAPG, KNL1, miR-148a-3p, miR-193b-3p, and miR-1179 may contribute to the progression of gastric cancer. Biol Res. 2018;51(1):44.
31. Jili S, Eryong L, Lijuan L, Chao Z. RUNX3 inhibits laryngeal squamous cell carcinoma malignancy under the regulation of miR-148a-3p/DNMT1 axis. Cell Biochem Funct. 2016;34(8):597–605.
32. Jacob H, Stanisavljevic L, Storli KE, Hestetun KE, Dahl O, Myklebust MP. Identification of a sixteen-microRNA signature as prognostic biomarker for stage II and III colon cancer. Oncotarget. 2017;8(50):87837–87847.
33. Wang W, Dong J, Wang M, Yao S, Zhang S. miR‑148a‑3p suppresses epithelial ovarian cancer progression primarily by targeting c‑Met. Oncol Lett. 2018;15(5):6131–6136.
34. Bhattacharya S, Chalk AM, Ng AJ, et al. Increased miR-155-5p and reduced miR-148a-3p contribute to the suppression of osteosarcoma cell death. Oncogene. 2016;35(40):5282–5294.
 © 2021 The Author(s). This work is published and licensed by Dove Medical Press Limited. The full terms of this license are available at https://www.dovepress.com/terms.php and incorporate the Creative Commons Attribution - Non Commercial (unported, v3.0) License.
By accessing the work you hereby accept the Terms. Non-commercial uses of the work are permitted without any further permission from Dove Medical Press Limited, provided the work is properly attributed. For permission for commercial use of this work, please see paragraphs 4.2 and 5 of our Terms.
© 2021 The Author(s). This work is published and licensed by Dove Medical Press Limited. The full terms of this license are available at https://www.dovepress.com/terms.php and incorporate the Creative Commons Attribution - Non Commercial (unported, v3.0) License.
By accessing the work you hereby accept the Terms. Non-commercial uses of the work are permitted without any further permission from Dove Medical Press Limited, provided the work is properly attributed. For permission for commercial use of this work, please see paragraphs 4.2 and 5 of our Terms.