Back to Journals » OncoTargets and Therapy » Volume 12
MicroRNA-145 Inhibits Cell Migration and Invasion in Colorectal Cancer by Targeting TWIST
Authors Shen X, Jiang H, Chen Z, Lu B, Zhu Y , Mao J, Chai K, Chen W
Received 17 May 2019
Accepted for publication 31 October 2019
Published 10 December 2019 Volume 2019:12 Pages 10799—10809
DOI https://doi.org/10.2147/OTT.S216147
Checked for plagiarism Yes
Review by Single anonymous peer review
Peer reviewer comments 2
Editor who approved publication: Dr Leo Jen-Liang Su
Xuning Shen,1 Honggang Jiang,1 Zhiheng Chen,1 Bohao Lu,1 Yi Zhu,1 Jiayan Mao,2 Kequn Chai,2,3 Wei Chen2,3
1Department of Gastroenterological Surgery, Affiliated Hospital of Jiaxing University, Jiaxing, Zhejiang, People’s Republic of China; 2Cancer Institute of Integrated Traditional Chinese and Western Medicine, Key Laboratory of Cancer Prevention and Therapy Combining Traditional Chinese and Western Medicine, Zhejiang Academy of Traditional Chinese Medicine, Hangzhou, Zhejiang 310012, People’s Republic of China; 3Department of Medical Oncology, Tongde Hospital of Zhejiang Province, Hangzhou, Zhejiang 310012, People’s Republic of China
Correspondence: Wei Chen; Kequn Chai
Cancer Institute of Integrated Traditional Chinese and Western Medicine, Key Laboratory of Cancer Prevention and Therapy Combining Traditional Chinese and Western Medicine, Zhejiang Academy of Traditional Chinese Medicine, Hangzhou, Zhejiang 310012, People’s Republic of China
Tel +86-571-89972114
Fax +86 571-88853199
Email [email protected]; [email protected]
Introduction: MicroRNAs function as oncogenes or tumor suppressors in the development of various human cancers. We investigated the effect of microRNA-145 (miR-145) on colorectal cancer (CRC) cell invasion and migration.
Methods: The levels of miR-145 in CRC cells were examined by quantitative PCR; Western blotting was used to detect TWIST1 (twist family bHLH transcription factor 1) protein and the epithelial–mesenchymal transition (EMT)-related proteins (E-cadherin, vimentin). Then, we transfected miR-145 mimics or inhibitor into CRC cells and used the wound healing and Transwell invasion assays to investigate their migration and invasive capability, respectively.
Results: The miR-145 mimics suppressed CRC cell invasion and migration significantly; in contrast, miR-145 downregulation had the opposite effect. Furthermore, miR-145 regulated TWIST1 levels negatively at transcriptional level. TWIST1 knockdown significantly inhibited the CRC cell migration ability and the number of CRC cells that crossed the Transwell membrane. There was no significant difference in terms of migration and invasive capability after the cells had been transfected with miR-145 mimics or inhibitor plus TWIST1 small interfering RNA (siRNA) as compared to the TWIST1 siRNA–only group. Furthermore, we demonstrate that the inhibition of miR-145 could enhance the capability for lung metastasis in vivo.
Conclusion: Taken together, these findings indicate that miR-145 acts as a new tumor suppressor by regulating TWIST1 and plays a vital role in the invasive and migration ability of CRC cells.
Keywords: MicroRNA-145, cell migration and invasion, colorectal cancer, TWIST, EMT
Introduction
Colorectal cancer (CRC) is the third most common malignant tumor in the world, with high tumor migration and invasion ability.1,2 In CRC, the development of normal epithelial cells into malignant tumors is considered to be a multi-stage process involving genetic alterations, leading to oncogene activation and tumor suppressor gene inactivation. Although migration and invasion are considered the deadliest features of solid tumors, the potential molecular mechanisms are rarely known. Therefore, it is very important to identify novel molecules with different expression in CRC cells, and it may aid the obtainment of insight into the involved mechanisms.
MicroRNAs (miRNAs) are small noncoding RNAs that are 18–22 nucleotides in length and that regulate gene expression negatively, affecting either mRNA cleavage or the translational inhibition of genes at post-transcriptional level.3 They play vital effects in tumorigenesis and metastasis, regulating gene expression through knockdown of mRNA translation, and also play oncogenic or tumor-suppressive roles according to the regulatory targets.4 Recent reports have indicated that miRNAs play important roles in the occurrence and development of different human cancers.5–7 miRNA-145 (miR-145), a tumor-suppressive miRNA, is downregulated in several malignancies such as breast cancer, lung cancer, bladder cancer, ovarian cancer, colon cancer, and prostate cancer.8–13 miR-145 was first found to be downregulated in CRC in 2003, and it is considered a tumor suppressor gene.14 Furthermore, miR-145 can regulate the expression response of a specific set of certain genes to tumorigenesis and metastasis by participating in CRC.14,15 However, the potential molecular mechanism of miR-145 in the progression of CRC is unclear.
TWIST1 (twist family bHLH transcription factor 1), a highly conserved basic helix-loop-helix (bHLH) transcription factor, is represented by a basic DNA-binding region targeting the consensus E-box sequence 5′-CANNTG-3′ and a HLH area.16 Epithelial–mesenchymal transition (EMT) is a new cellular process that is very important for the development of metastatic disease. TWIST1 is considered an inducer of EMT and a basic regulator of cancer metastasis.17,18 TWIST1 promotes cancer metastasis by promoting EMT.19.20 Based on its function as a cell adhesion molecule, the deletion of E-cadherin is considered a prerequisite for EMT to facilitate cancer cell proliferation and metastasis.19 Therefore, regulating the expression of TWIST1 might be a potential target for suppressing CRC cell invasion and migration.
In our present study, we investigated miR-145 and determined its effect on CRC cell invasion and migration. We prove that TWIST1 expression correlated negatively with miR-145 expression in CRC. We also observed that miR-145 overexpression inhibited the cell invasive and migration ability by regulating TWIST1 in CRC cells.
Materials and Methods
Cell Culture
Human CRC cells (LOVO, HCT116, HT29) were obtained from American Type Culture Collection (Manassas, VA, USA). LOVO and HT29 cells were maintained in RPMI 1640 medium (Gibco, Grand Island, NY, USA) supplemented with 10% fetal bovine serum (FBS; Gibco) and 1% penicillin/streptomycin (Sigma, St. Louis, MO, USA). The HCT116 cells were cultured in McCoy’s 5A medium (Gibco) supplemented with 10% FBS and 1% penicillin/streptomycin (Sigma). All cells were incubated at 37°C in a humidified atmosphere containing 5% CO2.
RNA Oligoribonucleotides and Transfection
miR-145 mimics, inhibitor, and negative control were synthesized by GenePharma (Shanghai, China). TWIST1 small interfering RNA (siRNA) was purchased from Santa Cruz Biotechnology (Santa Cruz, CA, USA). Lipofectamine 2000 (Invitrogen, Carlsbad, CA, USA) was used to transfect the CRC cells according to the manufacturer’s protocol. The oligoribonucleotide sequences were as follows:
miR-145 mimics: 5′-GUCCAGUUUUCCCAGGAAUCCCU-3′ and 5′-GGAUUCCUGGGAAAACUGGACUU-3′; miR-145 inhibitor: 5′-AGGGAUUCCUGGGAAAACUGGAC-3′; negative control: 5′-UUCUCCGAACGUGUCACGUTT-3′ and 5′-ACGUGACACGUUCGGAGAATT-3′.
Wound Healing Assay
The cell migration ability was determined using the wound healing assay. Cells were plated in 6-well plates at 3 × 105 cells/well, and then transfected with miR-145 mimics, inhibitor, or control. Yellow pipette tip was used to create a wound in the cell monolayer. After washing with phosphate-buffered saline, the wound area was photographed under an inverted light microscope (Olympus IX51, Olympus, Center Valley, PA, USA) at 0 h and 48 h. The ratio of residual wound area to initial wound area was calculated and the wound area was quantified by Image-Pro Plus v. 6.0 (Media Cybernetics, Bethesda, MD, USA).
Transwell Invasion Assay
Cell invasion was examined in 24-well Transwell chambers (8 μm; Corning, Inc.). First, we used Matrigel (10 μg/mL; BD Biosciences, San Jose, CA, USA) to cover the reverse side of the upper chambers and the surface was coated with 70 μL Matrigel (1 mg/mL). CRC cells (5 × 104) in 200 μl FBS-free medium were seeded in the upper chamber and 700 μL complete medium (with 10% FBS) was added to the bottom well. After 24-h incubation, cells that had invaded to the lower surfaces were gently cleaned and fixed with methanol for 10 min and stained with 0.1% crystal violet for 10 min. Five random fields were observed from the membrane, and the number of migrated cells was counted under an inverted phase contrast microscope (Olympus; ×40 magnification) and photographed.
Western Blotting
The cell proteins were extracted using cell lysis buffer (Cell Signaling Technology, Danvers, MA, USA), after which the protein concentration was determined by a bicinchoninic acid protein assay kit (BCA, Sigma-Aldrich; Merck KGaA). Protein samples (40 μg/lane) were separated by 10% sodium dodecyl sulphate-polyacrylamide gel electrophoresis (SDS-PAGE) and subsequently transferred to polyvinylidene difluoride (PVDF) membranes (Millipore, Billerica, MA, USA). The membranes were blocked with Tris-buffered saline and 0.1% Tween 20 (TBS-T) containing 5% bovine serum albumin, then incubated with primary antibody against E-cadherin, vimentin, and TWIST1 (diluted 1:1000 in TBS-T; Abcam, Cambridge, MA, USA). The membranes were incubated at 4°C overnight. After washing with TBS-T three times, the membranes were incubated with the appropriate horseradish peroxidase–labeled secondary antibody (diluted 1:2000 in TBS-T; Abcam) at 37°C for 2 h. GAPDH was used as the internal control. The proteins were examined using enhanced chemiluminescence reagents (Pierce, Rockford, IL, USA) and the band density was scanned using Image Lab 5.0 (Bio-Rad, Hercules, CA, USA).
Quantitative Real-Time PCR (qRT-PCR)
Total RNA was isolated using the TRIzol one-step method (Invitrogen). RNA (1 μg) was used, and complementary DNA (cDNA) was obtained by reverse transcription with a PrimeScript RT reagent kit according to the manufacturer’s instructions (Takara Biotechnology, Dalian, China). qPCR was performed on a 7500 Real-Time PCR system (Applied Biosystems, Foster City, CA, USA) using SYBR Premix Ex Taq II kits (Takara Biotechnology) according to the manufacturer’s instructions. The U6 RNA level was used as the internal control for miRNAs. Relative expression was calculated using the comparative threshold cycle (2−ΔΔCt) method.
Analysis of Lung Metastasis in Nude Mice
pCDH-CMV-luc-P2A-copGFP-T2A-Puro (Hangzhou Shiyu Biotechnology, Hangzhou, China) virus was transfected into a 6-well plate of LOVO cells. Stable strains were screened using 10 μg/mL puromycin, and observed under fluorescence microscopy. The cells were expanded to sufficient amounts, and then transfected with LV-miR-NC, LV-mimics-145, or LV-inhibitor-145. Female BALB/c nu/nu mice (5–6 weeks old) were randomly divided into four groups (n = 8 per group): normal saline, NC (negative control), miR-145, and miR-145 sponge. For lung metastasis analysis, 2 × 106 cells per mouse were injected into the tail vein of each nude mouse. The mice were weighed every other day, and changes in body weight were observed. After 4 weeks, the mice were injected intraperitoneally with D-luciferin. Then, they were anesthetized by isoflurane and photographed in an IVIS imaging system. The lungs were removed and fixed in 10% formalin. Subsequently, consecutive tissue sections were made from each lung block, and stained with hematoxylin–eosin (H&E). The experimental procedures and use of laboratory animals were approved by the Medical Ethics Committee of Tongde Hospital of Zhejiang Province, Hangzhou, China, and conformed to the National Institutes of Health Guide for Care and Use of Laboratory Animals (NIH Publications, No. 8023, revised 1978).
Statistical Analysis
All experimental data are reported as the mean ± standard deviation (n = 3); the data were analyzed using Prism 5 (GraphPad Software, La Jolla, CA, USA). Statistical differences between two groups were detected using the Student’s t-test; comparisons among multiple groups were performed using one-way analysis of variance, followed by Tukey’s post hoc test. P < 0.05 was considered to indicate a statistically significant difference.
Results
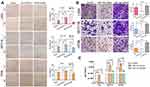
miR-145 Regulated CRC Cell Migration and Invasion
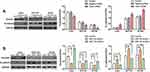
To explore whether miR-145 affects cell migration and invasion in CRC, we transfected the cells with miR-145 mimics or inhibitor, and then examined them using Transwell invasion and wound healing assays. miR-145 overexpression inhibited CRC cell migration ability, whereas miR-145 inhibitor enhanced it (Figure 1A and B). The Transwell invasion assay indicated that compared with the negative control, few cells crossed the membrane after miR-145 mimics transfection, but more cells crossed the membrane following miR-145 inhibitor transfection. qRT-PCR determined the interference efficiency of miR-145 following transfected with miR-145 mimic or inhibitor (Figure 1C). The results confirm that miR-145 can regulate CRC cell migration and invasive ability.
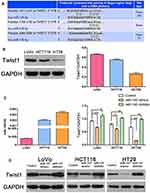
TWIST1 Was A Direct Target Gene of miR-145
We hypothesized that miR-145 regulates TWIST1. To verify this, we used TargetScan (www.targetscan.org) to predict whether TWIST1 is a target of miR-145 (Figure 2A), and the results were as we had expected. Next, we examined TWIST1 protein and miR-145 levels using Western blotting and qRT-PCR, respectively, and found that TWIST1 expression correlated negatively with miR-145 expression (Figure 2B and C). Then we transfected CRC cells with miR-145 mimics, inhibitor, or negative control and detected TWIST1 protein expression. miR-145 significantly downregulated TWIST1 levels, but the miR-145 inhibitor had the opposite effect (Figure 2D). These findings suggest that TWIST1 is a target gene of miR-145 in CRC cells.
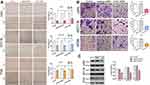
TWIST1 siRNA Reduced CRC Cell Migration and Invasive Capability
Increasing evidence suggests that TWIST1 is related with cell invasion and metastasis in various tumors, such as pancreatic cancer, ovarian cancer, and non–small cell lung cancer (NSCLC).21–23 To assess the role of TWIST1 in CRC cells, we transfected CRC cells with TWIST1 siRNA or negative siRNA, and determined the interference efficiency of TWIST1 siRNA using Western blotting (Figure 3C). The wound healing assay determined that, compared with negative siRNA, TWIST1 knockdown increased cell motility significantly and weakened CRC cell migration ability significantly (Figure 3A); the Transwell assay demonstrated significantly fewer invaded cells among the cells transfected with TWIST1 siRNA as compared with cells transfected with negative siRNA (Figure 3B), indicating that inhibiting TWIST1 suppresses CRC cell migration and invasive ability significantly.
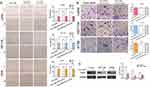
miR-145 Suppressed CRC Cell Migration and Invasion by Targeting TWIST
We demonstrated that miR-145 and TWIST1 can regulate cell invasion and migration and that miR-145 can regulate TWIST1 levels. We hypothesized that miR-145 regulates CRC cell invasion and migration by regulating TWIST1. To prove this, we silenced TWIST1 and then transfected CRC cells with miR-145 mimics or inhibitor. The results showed that there was no significant difference among the three groups, i.e., TWIST1 siRNA only, TWIST1 siRNA+miR-145 mimics, and TWIST1 siRNA+miR-145 inhibitor (Figure 4A and B). Western blotting confirmed TWIST1 expression following treatment with TWIST1 siRNA or negative control siRNA (Figure 4C).
miR-145 Overexpression Inhibited EMT in CRC Cells
Much evidence has indicated that EMT is associated with cell invasive and migration capability in different cancers, including hepatocellular carcinoma, pancreatic cancer, NSCLC, gastric cancer, and colon cancer.24–28 Here, we found that, compared with negative siRNA transfection, TWIST1 knockdown significantly upregulated E-cadherin expression and downregulated the level of vimentin in the CRC cells (Figure 5A). Furthermore, we found that miR-145 overexpression increased E-cadherin levels and decreased vimentin expression significantly, whereas miR-145 inhibition had the opposite effect (Figure 5B).Furthermore, we detected the expression of MMP-9 following transfection with miR-145 mimic or inhibitor in Figure S. These results indicate that miR-145 can regulate E-cadherin and vimentin levels in CRC cells.
miR-145 Regulated Metastasis in vivo
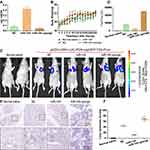
To further research the effect of miR-145 on CRC metastasis in vivo, we transfected LOVO cells with pCDH-CMV-LUC-P2A-copGFP-T2A-Puro and later transfected them with NC, miR-145 mimic, or miR-145 inhibitor to obverse lung metastasis in vivo. qRT-PCR determined the transfection efficiency of miR-145 treatment with miR-145 mimic or inhibitor (Figure 6A). Figure 6B shows the changes in body weight. Luciferase signals from the lungs of the miR-145 inhibitor group indicated higher metastasis ability compared with that of control group at 4 weeks post-injection (Figure 6C and D). H&E staining also showed that the capability for lung metastasis was faster following transfection with miR-145 inhibitor (Figure 6E and F). These results were consistent with the in vitro results.
Discussion
miRNAs can regulate 30% of human genes post-transcriptionally, suggesting that they may play a key role in physiological and pathological processes, including even human carcinogenesis.29 Recently, many reports have proven that miRNA expression is closely related to the occurrence, progression, metastasis, and prognosis of CRC.30,31 Accordingly, the identification of CRC-related miRNAs will facilitate understanding of the etiology of CRC and elucidate their biological effects in the diagnosis and prognosis. miR-145 can inhibit the migration and invasive ability in some cancer cells, such as breast cancer cells, glioma stem cells, prostate cancer cells, and NSCLC cells.8,32–34 In the present study, we prove that miR-145 overexpression inhibited CRC cell invasion and migration, while miR-145 inhibitor had the opposite effect. These findings indicate that miR-145 plays a vital role in regulating CRC cell invasion and migration.
TWIST1 is a novel oncogene overexpressed in a variety of tumors.35 Its activation inhibits apoptosis and promotes the induction of EMT and proliferation in human cancer cells, which contribute to metastasis.36,37 Much evidence has indicated that some miRNAs, such as miR-186, miR-151-3p, miR-720, miR-548c, miR-33a, and miR-520d-5p, directly regulate TWIST1 in a variety of cancer cells.38–43 miR-151-3p can repress invasion and metastasis in breast cancer by regulating TWIST1 expression.39 Here, we explored the role of the molecular mechanism of miR-145. TargetScan identified TWIST1 as a direct functional downstream target of miR-145. Moreover, miR-145 expression was negatively associated with TWIST1 expression. TWIST1 knockdown inhibited CRC cell invasion and migration. To explore whether miR-145 actually suppressed CRC cell migration and invasion by targeting TWIST1, we silenced TWIST1, then transfected CRC cells with miR-145 mimics and inhibitor, found no significant difference in cell migration and invasion compared with that of cells transfected with TWIST1 siRNA alone. These results indicate that the role of miR-145 in inhibiting CRC cell invasion and migration is associated with TWIST1 expression.
EMT is a complex transdifferentiation process that is accompanied by the loss of junctional adhesion and changes in mesenchymal phenotype and morphology, resulting in increased cell invasion and migration.44 EMT activation causes tumor cell invasion and spread, so it is considered the first step of metastasis.44 TWIST1 is a well-known transcription factor in EMT progression. Recently, it was reported that most miRNAs participated in tumor metastasis by targeting important EMT genes.45 Liu et al and You et al have revealed that the overexpression of miR-132 or miR-204 suppresses lung cancer cell migration and invasion by targeting EMT.27,46 miR-145 can also suppress breast cancer cell migration by targeting and inhibiting EMT.8 Furthermore, others have reported that miRNAs can regulate cell invasion and migration by targeting TWIST1 and inhibiting EMT in cancer cells.39,47 In the present study, we determined that miR-145 is involved in cell invasion and migration by regulating TWIST1 and EMT. Western blotting indicated that transfection with miR-145 mimics significantly upregulated the level of E-cadherin and downregulated vimentin expression, compared with the negative control, but the miR-145 inhibitor had the opposite effect. Moreover, the increased expression of matrix metalloproteinases (MMPs) is associated with increased potential for metastasis.48 Therefore, we determined the expression of MMP-9 following transfection with miR-145 mimic or inhibitor, and show that miR-145 mimic downregulated MMP-9, while miR-145 inhibitor upregulated it (Figure S1).
In conclusion, we demonstrate that the overexpression of miR-145 can inhibit CRC cell invasion and migration by targeting TWIST1 and inhibiting EMT. These findings indicate that miR-145 plays an important role in regulating CRC cell invasion and migration.
Acknowledgements
This study was funded by Zhejiang Basic Public Welfare Research Project (No. LGF18H160033), Zhejiang Provincial Natural Science Foundation of China (No. LQ13H160006), 2019 Jixing Key Discipline of Medicine-Oncology (Supporting Subject; 2019-zc-11), the Medical and Health Science and Technology Project of Zhejiang Province, China (2019KY214), Zhejiang Provincial Ten Thousand Plan for Young Top Talents (2018), Training objects of health innovative talents of Zhejiang Health (2018), Key Project Co-constructed by Zhejiang Province and Ministry (WKJ-ZJ-1916) and Natural Science Foundation of China (81972693, 81972674 and 31900543). Kequn Chai and Wei Chen are co-corresponding authors.
Disclosure
The authors report no conflicts of interest in this work.
References
1. Weitz J, Koch M, Debus J, Hohler T, Galle PR, Buchler MW. Colorectal cancer. Lancet. 2005;365(9454):153–165. doi:10.1016/S0140-6736(05)17706-X
2. Haggar FA, Boushey RP. Colorectal cancer epidemiology: incidence, mortality, survival, and risk factors. Clin Colon Rectal Surg. 2009;22(4):191–197. doi:10.1055/s-0029-1242458
3. Bartel DP. MicroRNAs: genomics, biogenesis, mechanism, and function. Cell. 2004;116(2):281–297. doi:10.1016/S0092-8674(04)00045-5
4. Shenouda SK, Alahari SK. MicroRNA function in cancer: oncogene or a tumor suppressor? Cancer Metastasis Rev. 2009;28(3–4):369–378. doi:10.1007/s10555-009-9188-5
5. Liu CG, Calin GA, Meloon B, et al. An oligonucleotide microchip for genome-wide microRNA profiling in human and mouse tissues. Proc Natl Acad Sci U S A. 2004;101(26):9740–9744. doi:10.1073/pnas.0403293101
6. Ambros V. The functions of animal microRNAs. Nature. 2004;431(7006):350–355.
7. Volinia S, Calin GA, Liu CG, et al. A microRNA expression signature of human solid tumors defines cancer gene targets. Proc Natl Acad Sci U S A. 2006;103(7):2257–2261. doi:10.1073/pnas.0510565103
8. Zhao H, Kang X, Xia X, et al. miR-145 suppresses breast cancer cell migration by targeting FSCN-1 and inhibiting epithelial-mesenchymal transition. Am J Transl Res. 2016;8(7):3106–3114.
9. Hu H, Xu Z, Li C, et al. MiR-145 and miR-203 represses TGF-beta-induced epithelial-mesenchymal transition and invasion by inhibiting SMAD3 in non-small cell lung cancer cells. Lung Cancer. 2016;97:87–94. doi:10.1016/j.lungcan.2016.04.017
10. Xue M, Pang H, Li X, Li H, Pan J, Chen W. Long non-coding RNA urothelial cancer-associated 1 promotes bladder cancer cell migration and invasion by way of the hsa-miR-145-ZEB1/2-FSCN1 pathway. Cancer Sci. 2016;107(1):18–27. doi:10.1111/cas.12844
11. Dong R, Liu X, Zhang Q, et al. miR-145 inhibits tumor growth and metastasis by targeting metadherin in high-grade serous ovarian carcinoma. Oncotarget. 2014;5(21):10816–10829. doi:10.18632/oncotarget.v5i21
12. Xu X, Wu X, Jiang Q, et al. Downregulation of microRNA-1 and microRNA-145 contributes synergistically to the development of colon cancer. Int J Mol Med. 2015;36(6):1630–1638. doi:10.3892/ijmm.2015.2364
13. Larne O, Hagman Z, Lilja H, Bjartell A, Edsjo A, Ceder Y. miR-145 suppress the androgen receptor in prostate cancer cells and correlates to prostate cancer prognosis. Carcinogenesis. 2015;36(8):858–866. doi:10.1093/carcin/bgv063
14. Michael MZ, O’ Connor SM, van Holst Pellekaan NG, Young GP, James RJ. Reduced accumulation of specific microRNAs in colorectal neoplasia. Mol Cancer Res. 2003;1(12):882–891.
15. Feng Y, Zhu J, Ou C, et al. MicroRNA-145 inhibits tumour growth and metastasis in colorectal cancer by targeting fascin-1. Br J Cancer. 2014;110(9):2300–2309. doi:10.1038/bjc.2014.122
16. Wushou A, Hou J, Zhao YJ, Shao ZM. Twist-1 up-regulation in carcinoma correlates to poor survival. Int J Mol Sci. 2014;15(12):21621–21630. doi:10.3390/ijms151221621
17. Rosivatz E, Becker I, Specht K, et al. Differential expression of the epithelial-mesenchymal transition regulators snail, SIP1, and twist in gastric cancer. Am J Pathol. 2002;161(5):1881–1891. doi:10.1016/S0002-9440(10)64464-1
18. Parmar MK, Torri V, Stewart L. Extracting summary statistics to perform meta-analyses of the published literature for survival endpoints. Stat Med. 1998;17(24):2815–2834. doi:10.1002/(SICI)1097-0258(19981230)17:24<2815::AID-SIM110>3.0.CO;2-8
19. Kalluri R, Weinberg RA. The basics of epithelial-mesenchymal transition. J Clin Invest. 2009;119(6):1420–1428. doi:10.1172/JCI39104
20. Chou YS, Yang MH. Epithelial-mesenchymal transition-related factors in solid tumor and hematological malignancy. J Chin Med Assoc. 2015;78(8):438–445. doi:10.1016/j.jcma.2015.05.002
21. Ji H, Lu HW, Li YM, et al. Twist promotes invasion and cisplatin resistance in pancreatic cancer cells through growth differentiation factor 15. Mol Med Rep. 2015;12(3):3841–3848. doi:10.3892/mmr.2015.3867
22. Wang WS, Yang XS, Xia M, Jiang HY, Hou JQ. Silencing of twist expression by RNA interference suppresses epithelial-mesenchymal transition, invasion, and metastasis of ovarian cancer. Asian Pac J Cancer Prev. 2012;13(9):4435–4439. doi:10.7314/APJCP.2012.13.9.4435
23. Wei L, Sun JJ, Cui YC, et al. Twist may be associated with invasion and metastasis of hypoxic NSCLC cells. Tumour Biol. 2016;37(7):9979–9987. doi:10.1007/s13277-016-4896-2
24. Lin Z, Li W, Zhang H, et al. CCL18/PITPNM3 enhances migration, invasion, and EMT through the NF-kappaB signaling pathway in hepatocellular carcinoma. Tumour Biol. 2016;37(3):3461–3468. doi:10.1007/s13277-015-4172-x
25. Zhang Z, Che X, Yang N, et al. miR-135b-5p promotes migration, invasion and EMT of pancreatic cancer cells by targeting NR3C2. Biomed Pharmacother. 2017;96:1341–1348. doi:10.1016/j.biopha.2017.11.074
26. Yue QY, Zhang Y. Effects of Linc00460 on cell migration and invasion through regulating epithelial-mesenchymal transition (EMT) in non-small cell lung cancer. Eur Rev Med Pharmacol Sci. 2018;22(4):1003–1010. doi:10.26355/eurrev_201802_14382
27. Liu Z, Long J, Du R, Ge C, Guo K, Xu Y. miR-204 regulates the EMT by targeting snai1 to suppress the invasion and migration of gastric cancer. Tumour Biol. 2016;37(6):8327–8335. doi:10.1007/s13277-015-4627-0
28. Dinicola S, Masiello MG, Proietti S, et al. Nicotine increases colon cancer cell migration and invasion through epithelial to mesenchymal transition (EMT): COX-2 involvement. J Cell Physiol. 2018;233(6):4935–4948. doi:10.1002/jcp.v233.6
29. Lewis BP, Burge CB, Bartel DP. Conserved seed pairing, often flanked by adenosines, indicates that thousands of human genes are microRNA targets. Cell. 2005;120(1):15–20. doi:10.1016/j.cell.2004.12.035
30. de Krijger I, Mekenkamp LJ, Punt CJ, Nagtegaal ID. MicroRNAs in colorectal cancer metastasis. J Pathol. 2011;224(4):438–447. doi:10.1002/path.2922
31. Muhammad S, Kaur K, Huang R, et al. MicroRNAs in colorectal cancer: role in metastasis and clinical perspectives. World J Gastroenterol. 2014;20(45):17011–17019. doi:10.3748/wjg.v20.i45.17011
32. Lu Y, Chopp M, Zheng X, Katakowski M, Buller B, Jiang F. MiR-145 reduces ADAM17 expression and inhibits in vitro migration and invasion of glioma cells. Oncol Rep. 2013;29(1):67–72. doi:10.3892/or.2012.2084
33. Xie S, Xie Y, Zhang Y, Huang Q. [Effects of miR-145 on the migration and invasion of prostate cancer PC3 cells by targeting DAB2]. Yi Chuan. 2014;36(1):50–57. doi:10.3724/SP.J.1005.2014.00050
34. Zhang Y, Yang X, Wu H, Zhou W, Liu Z. MicroRNA-145 inhibits migration and invasion via inhibition of fascin 1 protein expression in non-small-cell lung cancer cells. Mol Med Rep. 2015;12(4):6193–6198. doi:10.3892/mmr.2015.4163
35. Kang Y, Massague J. Epithelial-mesenchymal transitions: twist in development and metastasis. Cell. 2004;118(3):277–279. doi:10.1016/j.cell.2004.07.011
36. Khan MA, Chen HC, Zhang D, Fu J. Twist: a molecular target in cancer therapeutics. Tumour Biol. 2013;34(5):2497–2506. doi:10.1007/s13277-013-1002-x
37. Ma L, Teruya-Feldstein J, Weinberg RA. Tumour invasion and metastasis initiated by microRNA-10b in breast cancer. Nature. 2007;449(7163):682–688. doi:10.1038/nature06174
38. Cao C, Sun D, Zhang L, Song L. miR-186 affects the proliferation, invasion and migration of human gastric cancer by inhibition of Twist1. Oncotarget. 2016;7(48):79956–79963. doi:10.18632/oncotarget.v7i48
39. Yeh TC, Huang TT, Yeh TS, et al. miR-151-3p targets TWIST1 to repress migration of human breast cancer cells. PLoS One. 2016;11(12):e0168171. doi:10.1371/journal.pone.0168171
40. Li LZ, Zhang CZ, Liu LL, et al. miR-720 inhibits tumor invasion and migration in breast cancer by targeting TWIST1. Carcinogenesis. 2014;35(2):469–478. doi:10.1093/carcin/bgt330
41. Sun X, Cui M, Zhang A, et al. MiR-548c impairs migration and invasion of endometrial and ovarian cancer cells via downregulation of twist. J Exp Clin Cancer Res. 2016;35:10. doi:10.1186/s13046-016-0288-0
42. Zhou Y, Huang Z, Wu S, Zang X, Liu M, Shi J. miR-33a is up-regulated in chemoresistant osteosarcoma and promotes osteosarcoma cell resistance to cisplatin by down-regulating TWIST. J Exp Clin Cancer Res. 2014;33:12. doi:10.1186/1756-9966-33-12
43. Tsukerman P, Yamin R, Seidel E, et al. MiR-520d-5p directly targets TWIST1 and downregulates the metastamiR miR-10b. Oncotarget. 2014;5(23):12141–12150. doi:10.18632/oncotarget.v5i23
44. Thiery JP. Epithelial-mesenchymal transitions in tumour progression. Nat Rev Cancer. 2002;2(6):442–454. doi:10.1038/nrc822
45. Ding XM. MicroRNAs: regulators of cancer metastasis and epithelial-mesenchymal transition (EMT). Chin J Cancer. 2014;33(3):140–147. doi:10.5732/cjc.013.10094
46. You J, Li Y, Fang N, et al. MiR-132 suppresses the migration and invasion of lung cancer cells via targeting the EMT regulator ZEB2. PLoS One. 2014;9(3):e91827. doi:10.1371/journal.pone.0091827
47. Yu J, Xie F, Bao X, Chen W, Xu Q. miR-300 inhibits epithelial to mesenchymal transition and metastasis by targeting Twist in human epithelial cancer. Mol Cancer. 2014;13:121.
48. Shih YW, Shieh JM, Wu PF, Lee YC, Chen YZ, Chiang TA. Alpha-tomatine inactivates PI3K/Akt and ERK signaling pathways in human lung adenocarcinoma A549 cells: effect on metastasis. Food Chem Toxicol. 2009;47(8):1985–1995. doi:10.1016/j.fct.2009.05.011
 © 2019 The Author(s). This work is published and licensed by Dove Medical Press Limited. The full terms of this license are available at https://www.dovepress.com/terms.php and incorporate the Creative Commons Attribution - Non Commercial (unported, v3.0) License.
By accessing the work you hereby accept the Terms. Non-commercial uses of the work are permitted without any further permission from Dove Medical Press Limited, provided the work is properly attributed. For permission for commercial use of this work, please see paragraphs 4.2 and 5 of our Terms.
© 2019 The Author(s). This work is published and licensed by Dove Medical Press Limited. The full terms of this license are available at https://www.dovepress.com/terms.php and incorporate the Creative Commons Attribution - Non Commercial (unported, v3.0) License.
By accessing the work you hereby accept the Terms. Non-commercial uses of the work are permitted without any further permission from Dove Medical Press Limited, provided the work is properly attributed. For permission for commercial use of this work, please see paragraphs 4.2 and 5 of our Terms.