Back to Journals » OncoTargets and Therapy » Volume 12
MicroRNA-139-3p Inhibits The Growth And Metastasis Of Ovarian Cancer By Inhibiting ELAVL1
Authors Xue F, Li QR, Xu YH, Zhou HB
Received 1 April 2019
Accepted for publication 3 September 2019
Published 5 November 2019 Volume 2019:12 Pages 8935—8945
DOI https://doi.org/10.2147/OTT.S210739
Checked for plagiarism Yes
Review by Single anonymous peer review
Peer reviewer comments 2
Editor who approved publication: Dr Sanjeev K. Srivastava
This paper has been retracted.
Fang Xue,1 Qi Rong Li,1 Yan Hua Xu,1 Hai Bin Zhou2
1Department of Gynaecology and Obstetrics, Jinan Maternal and Child Health Hospital Jinan, Shandong 250012, People’s Republic of China; 2Infertility Center, Department of Gynaecology and Obstetrics, Qilu Hospital of Shandong University, Jinan, Shandong 250012, People’s Republic of China
Correspondence: Hai Bin Zhou
Infertility Center, Department of Gynaecology and Obstetrics, Qilu Hospital of Shandong University, Jinan, Shandong 250012, People’s Republic of China
Email [email protected]
Background: The aberrant expression of microRNA-139-3p (miR-139-3p) has been recently involved in the development of multiple tumor types, but its function in ovarian cancer remains not well investigated. In this study, we mainly investigated the function of miR-139-3p in the progression of ovarian cancer.
Methods: The levels of miR-139-3p in ovarian cancer cells and tissues were detected using quantitative real-time-PCR (qRT-PCR) assay. The proliferation, colony formation, migration and invasion of ovarian cancer cell were determined, respectively. A luciferase reporter assay was used to confirm ELAV Like RNA Binding Protein 1 (ELAVL1) was a target gene of miR-139-3p. The expression of ELAVL1 was detected using Western blotting and immunofluorescence staining assay. The roles of miR-139-3p on the growth and metastasis of ovarian cancer cell in vivo were explored using transplanted tumor model and experimental lung metastasis model.
Results: MiR-139-3p was down-regulated in ovarian cancer tissues and ovarian cancer cell lines (SK-OV-3, A2780 and OVCAR-3). Overexpression of miR-139-3p decreased the growth, colony formation, migration and invasiveness of SK-OV-3 and OVCAR-3 cells. Moreover, overexpression of miR-139-3p reduced the growth and lung metastasis of ovarian cancer cells in vivo. The luciferase reporter gene assay indicated that ELAVL1 was a target of miR-139-3p and its expression was negatively regulated by miR-139-3p. Furthermore, the expression of ELAVL1 was inversely correlated with miR-139-3p level in ovarian cancer tissue.
Conclusion: Taken together, we demonstrated that miR-139-3p regulated ovarian cancer growth and metastasis by modulating the expression of ELAVL1.
Keywords: ovarian cancer, MiR-139-3p, ELAVL1, metastasis, growth
Background
Ovarian cancer is one of the most deadly malignant tumors in gynecologic oncology, characterized by the poor prognosis due to the extensive metastasis of ovarian cancer cell into the peritoneal cavity.1 Despite the extensive basic and clinical investigations, only 27% of patients with advanced ovarian carcinoma survived for 5 years after the initial diagnosis.2 There is an urgent need to explore the underlying mechanism of ovarian carcinoma metastasis and improve the targeted therapeutic strategies.3
MicroRNAs (miRNAs), which are a kind of conserved non-coding RNAs, regulate the expression of proteins via binding with the 3ʹ-untranslated region (3ʹ-UTR) of target mRNA.4,5 The functions of miRNAs in many biological processes, including cancer cell growth, apoptosis, migration and metastasis, have been widely investigated.6 In oral squamous carcinoma cell (OSCC), miR-139-5p suppresses the tumorigenesis and progression of OSCC via targeting Homeobox A9 (HOXA9).7 In addition, miR-139-5p is markedly down-regulated in uterine leiomyoma tissue than that in the adjacent myometrium tissue and miR-139-5p inhibits the growth of uterine leiomyoma cells through regulating Tumor Protein D52 (TPD52).8 All these investigations indicate that miR-139-5p acts as a tumor-suppressive role in several cancers.9–11 Recently, the impacts of miRNAs in the progression of ovarian cancer have already been explored. Nevertheless, the precise function of miR-139-3p in ovarian cancer remains largely unknown.
Herein, we revealed that miRNA-139-3p was down-expressed in ovarian cancer tissues and cells. The roles of miRNA-139-3p on the development of ovarian cancer cell in vitro and in vitro were analyzed. Importantly, we validated that ELAVL1 was a functional target gene of miRNA-139-3p and its expression was negatively correlated with the level of miRNA-139-3p in ovarian cancer. Finally, we proved that over-expression of miRNA-139-3p suppressed the growth and aggressiveness of ovarian cancer through regulating the expression of ELAVL1.
Materials And Methods
Clinical Tissues
Twenty-one cases of ovarian cancer and adjacent normal tissues were obtained from the Qilu Hospital of Shandong University from 2007 to 2017. No patients with ovarian cancer underwent chemotherapy and radiotherapy before sample biopsy. Written consent was obtained from all patients who were involved in the study. The clinicopathological parameters in patients with ovarian cancer were summarized in Supplementary Table 1. Ethical approval was obtained from the Ethics Committee of the Qilu Hospital of Shandong University. The study conforms to the Code of Ethics of the World Medical Association (Declaration of Helsinki) printed in the British Medical Journal (18 July 1964).
Cell Culture
Human ovarian carcinoma cells (OVCAR-3, SK-OV-3 and A2780) and normal ovarian epithelial cell, IOSE80, were purchased from Chinese Academy of Sciences (Shanghai, China). Cells were cultured using RPMI media 1640 (Thermo Fisher Scientific, Waltham, MA, USA) containing 10% FBS (Thermo Fisher Scientific) and 1% streptomycin/penicillin at 37°C and 5% CO2.
Cell Transfection
MiRNA-negative control (miR-NC) or miR-139-3p was purchased from GeneCopoeia (Shanghai, China). A ELAVL1 expression was constructed by subcloning PCR-amplified full-length ELAVL1 cDNA into pcDNA3.1(+) plasmid (GeneCopoeia). MiR-139-3p or pCDNA3.1-ELAVL1 was transfected into cell using Lipofectamine™ 2000 (Thermo Fisher Scientific).
Quantitative Real-Time PCR (qRT-PCR)
Total RNA was extracted using TRIzol reagent (Thermo Fisher Scientific). The level of miRNA-139-3p was detected using a SYBR PrimeScript miRNA RT PCR kit (Takara, Dalian, China). For detection of the mRNA level of ELAVL1, first-strand cDNA synthesis was conducted using MMLV reverse transcriptase (Promega). qRT-PCR was conducted using SYBR Premix Ex Taq (TaKaRa) in ABI 7900 Fast system (Applied Biosystems, CA, USA). The primers for ELAVL1 and GAPDH were as following: ELAVL1 forward: 5ʹ-ATGAAGACCACATGGCCGAAGACT-3ʹ; reverse: 5ʹ-AGTTCACAAAGCCATAGCCCAAGC-3ʹ; GAPDH forward: 5′-ACAACTTTGGTATCGTGGAAGG-3′; reverse: 5′-GCCATCACGCCACAGTTTC-3′. miR-129-3p forward: 5′-AAGCCCTTACCCCAAAAAGTAT-3′; reverse: 5′-CTTTTTGCGGTCTGGGCTTGC-3′; fibronection forward: 5′-CGGTGGCTGTCAGTCAAAG-3′; reverse: 5′-AAACCTCGGCTTCCTCCATAA-3′; Vimentin forward: 5′-GACGCCATCAACACCGAGTT-3′; reverse: 5′-CTTTGTCGTTGGTTAGCTGGT-3′; ZO-1 forward: 5′-CAACATACAGTGACGCTTCACA-3′; reverse: 5′-CACTATTGACGTTTCCCCACTC-3′; β-catenin forward: 5′-AAAGCGGCTGTTAGTCACTGG-3′; reverse: 5′-CGAGTCATTGCATACTGTCCAT-3′; U6 forward: 5ʹ-CTCGCTTCGGCAGCACA-3ʹ, U6 reverse: 5ʹ-AACGCTTCACGAATTTGCGT-3ʹ. The level of miR-139-3p and ELAVL1 were normalized to U6 or GAPDH, respectively, using the 2−ΔΔCT method.12
CCK-8 Assay
Cells (2 × 103) were cultured into 96-well plates and were cultured for 24 hrs, 48 hrs, 73 hrs or 96 hrs. Then, 10 μL of CCK-8 solution (Beyotime, Nanjing, Jiangsu, China) was added into plates and cells were incubated for 1.5 hrs. The OD was detected at 450 nm using a microplate reader (Bio-Tek Company, Winooski, VT, USA).
Colony Formation Analysis
Cells (1 × 103) were seeded into 6-well plates and cultured for two weeks. After that, cell colonies were fixed using 10% paraformaldehyde and stained using 1% crystal violet for 15 mins. Finally, the number of cell colonies was counted.
Migration Assay
Cells (1×105/well) were cultured into 6-well plates for 24 hrs. Then, a wound was made using a micropipette tip. The cells were cultured with FBS-free medium for 24 hrs. The pictures of wound were taken at 0 hrs or 24 hrs under an inverted microscope (CarlZeiss, Hallbergnoos, Germany).
Invasion Assay
200 μL cells (2×105) in serum-free media were cultured into the upper chamber of the transwell membrane (Transwell Permeable Support with an 8.0 μm polycarbonate membrane, 6.5 mm insert and 24-well plate; Corning Costar, New York, NY, USA). 600 μL medium containing 20% FBS was added into the lower chamber. After 24 hrs, invaded cells were fixed using 70% ethanol and stained with 1% crystal violet. The invaded cells were counted under an inverted microscope (CarlZeiss).
Luciferase Reporter Gene Analysis
The wild-type (wt) 3ʹ-UTR of ELAVL1 containing the predicted miR-139-3p-binding site was PCR amplified using PCR and inserted into the psiCHECK2 vector (Promega, Wisconsin, WI, USA). The mutant type (mut) 3ʹ-UTR of ELAVL1 was constructed using QuikChange XL Site-Directed Mutagenesis Kit (Agilent Technologies; Santa Clara, CA, USA) and inserted into psiCHECK2 vector. MiR-139-3p or miR-NC in combination with wt-ELAVL1-3ʹ-UTR or mut-ELAVL1-3ʹ-UTR was cotransfected into 293T cell using Lipofectamine™ 2000 (Thermo Fisher Scientific). The luciferase activity was detected using the luciferase reporter gene assay system (Promega).
Western Blotting
Total proteins were isolated from cells using RIPA buffer (Beyotime, Shanghai, China) on ice. The protein concentration was quantified using a BCA Protein Assay Kit (Beyotime). 30 μg proteins were fractionated using 8% SDS-PAGE and were transferred onto PVDF. The PVDF membrane was incubated with ELAVL1 antibody (#12,582, Cell Signaling Technology) or GAPDH (#5174, Cell Signaling Technology, Danvers, MA, USA) followed by incubation with HRP-linked secondary antibody (#7074, Cell Signaling Technology). The bands were assessed using ECL system (Millipore, Braunschweig, Germany).
In Vivo Xenograft Model
MiR-139-3p or miR-NC-transfected SK-OV-3 cells (2×106) were subcutaneously injected into BALB/c nu/nu mice (n=6 in each group). Tumor tissue was monitored each week using a digital caliper by measuring the tumor length (L) and width (W). The tumor volume was calculated as (L × W2)/2. After 35 days, nude mice were sacrificed, and the tumor tissue was weighed. Animal experiment was approved by the Institutional Animal Care and Use Committee (IACUC) of Qilu Hospital of Shandong University. The animal experiment was conducted in accordance with Institutional Guidelines and the Guide for the Care and Use of Laboratory Animals (NIH publication no. 85–23, revised 1996).
Experimental Lung Metastasis Assay
5×105 SK-OV-3 cells were injected into nude mice via the lateral tail vein. Mice were sacrificed after two weeks and lung tissues were used for IHC staining using ELAVL1 antibody. The metastatic nodules in lung were counted under a dissecting microscope. Animal experiment was approved by the Institutional Animal Care and Use Committee (IACUC) of Qilu Hospital of Shandong University. The animal experiment was conducted in accordance with Institutional Guidelines and the Guide for the Care and Use of Laboratory Animals (NIH publication no. 85–23, revised 1996).
Statistical Analysis
Data were obtained from at least three independent experiments and were presented as means ± SD. Data from all experimental groups were analyzed using one-way analysis of variance (ANOVA), followed by the LSD post hoc test using SPSS v. 21.0 (SPSS Inc., Chicago, IL, USA). The relationship between miR-139-3p and ELAVL1 was detected using Spearman correlation analysis. P<0.05 was considered statistically significant.
Results
miR-139-3p Is Down-Expressed In Ovarian Cancer
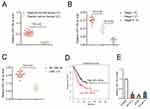
The qRT-PCR result indicated that miR-139-3p was markedly down-expressed in ovarian cancer tissues than that in corresponding normal tissues (*P<0.05, Figure 1A). Moreover, the level of miR-139-3p was significantly decreased in advanced stage of ovarian cancer than that in low stage (*P<0.05, Figure 1B). Meanwhile, miR-139-3p was down-expressed in patient with lymph node metastasis (LNM) than that in patient without LNM (No LNM) (Figure 1C). Next, we determined the prognostic significance of miR-129-3p in ovarian cancer. We divided patients into miR-139-3p low and miR-139-3p high groups using the median value of 0.36 as a cut-off value. The Kaplan–Meier survival analysis indicated that patients with lower level of miR-139-3p had worse survival than patients with higher level of miR-139-3p (Figure 1D). Finally, we also found that miR-139-3p was down-regulated in ovarian cancer cells, including SK-OV-3, A2780 and OVCAR-3, than that in the normal ovarian epithelial cell, IOSE80 (Figure 1E). We also detected the expression of miR-139-5p in ovarian cancer tissues and ovarian cancer cell lines. As shown in Supplementary Figure 1A, there was no significant difference in miR-139-5p between ovarian cancer tissues and corresponding normal tissues. Consistently, miR-139-5p was not remarkably downregulated in ovarian cancer cell lines than that in ovarian epithelial cell, IOSE80 (Supplementary Figure 1A). The result suggests that miR-139-3p is down-regulated in ovarian cancer tissues and cell lines.
Up-Regulation Of miR-139-3p Inhibits Ovarian Cancer Cell Growth
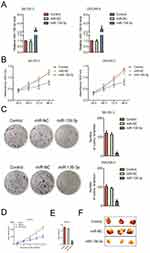
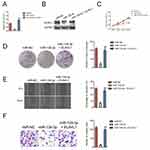
To investigate the biological function of miR-139-3p in ovarian carcinoma, miR-NC or miR-139-3p was transfected into OVCAR-3 and SK-OV-3 cell and the level of miR-139-3p was assessed by qRT-PCR test (Figure 2A). Then, the CCK-8 analysis demonstrated that up-regulation of miR-139-3p reduced the proliferation of SK-OV-3 and OVCAR-3 cells (Figure 2B). Moreover, miR-139-3p transfection inhibited the colony formation of SK-OV-3 and OVCAR-3 cells in vitro (Figure 2C). The similar results were obtained using A2780 cell line (Supplementary Figure 2). To future confirm the findings in vivo, miR-139-3p or miR-NC-transfected SK-OV-3 cell was subcutaneously injected into nude mice. We observed that the tumor growth of SK-OV-3 cells in the miR-139-3p group was significantly inhibited than that in the miR-NC group (Figure 2D–E). After 35 days, we observed that the tumor size in miR-139-3p group was lower than that in the miR-NC group (Figure 2F). All these results demonstrated that miR-139-3p inhibited the proliferation of ovarian cancer cell in vitro and tumor growth of SK-OV-3 cell in vivo.
miR-139-3p Suppresses The Migration And Invasion Of Ovarian Cancer Cell
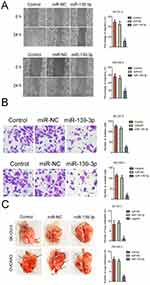
Next, we explored the function of miR-139-3p in the aggressiveness of ovarian cancer cell in vitro. We found that miR-139-3p significantly inhibited the migration and invasion of SK-OV-3 and OVCAR-3 cells (Figure 3A–B). To further investigate the role of miR-139-3p in the metastasis of SK-OV-3 cells in vivo, the experimental lung metastasis model was constructed. MiR-139-3p-transfected SK-OV-3 or OVCAR3 cell was injected into nude mice via tail vein. After 2 weeks, mice were sacrificed and metastatic nodules were mainly found on the lung tissue. As shown in Figure 3C, up-regulation of miR-139-3p significantly suppressed pulmonary metastasis. To understand the inhibitory effect of miR-139-3p on ovarian cancer cell epithelial-mesenchymal transition (EMT), Western blotting and qRT-PCR assay was applied to detect the different expressions of E-cadherin, N-cadherin, in SK-OV-3 and OVCRA3 cell. As shown in Supplementary Figure 3, transfection of miR-139-3p inhibited the expression of mesenchymal markers (N-cadherin, fibronection and Vimentin) and increased the levels of epithelial-related markers (E-cadherin, ZO-1 and β-catenin). These results implied that over-expression of miR-139-3p reduced the metastasis of SK-OV-3 cell in vivo.
ELAVL1 Is The Target Of miR-139-3p
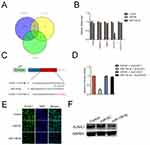
Next, we predicted the potential targets of miR-139-3p using three tools (TargetScan, miRTarBase and miRDB). As shown in Figure 4A, we found 5 common elements (FN3K, UBE2G1, MIEF1, CCDC71L and ELAVL1) in “TargetScan”, “miRTarBase” and “miRDB”. In order to identify the direct target of miR-139-3p, the levels of these genes in miR-NC or miR-139-3p transfected SK-OV-3 cell were detected using qRT-PCR assay. As shown in Figure 4B, the level of ELAVL1 (HuR) was markedly reduced by miR-139-3p.; luciferase reporter gene assay was used to identify ELAVL1 as the potential target gene of miR-139-3p (Figure 4C–D). The luciferase activity in 293T cell that was cotransfected with miR-139-3p and wt-ELAVL1 was decreased whereas the luciferase activity in cell that was cotransfected with miR-139-3p and mut-ELAVL1 was not significantly inhibited by miR-139-3p. Moreover, over-expression of miR-139-3p decreased the expression of ELAVL1 in ovarian cancer SK-OV-3 cell (Figure 4E–F). Finally, the immunohistochemistry (IHC) staining also indicated that miR-139-3p led to a marked reduction of ELAVL1 expression in lung metastasis foci formed by miR-139-3p transfection ovarian cancer cell (Supplementary Figure 4). All these results indicated that ELAVL1 is a direct target gene of miR-139-3p and negatively regulated by miR-139-3p.
ELAVL1 Mediates The Functional Effects Of miR-139-3p On Ovarian Cancer Cell
Then, SK-OV-3 cells were transfected with miR-139-3p mimics and ELAVL1 overexpressing plasmid to restore the expression of ELAVL1 levels in SK-OV-3 cells (Figure 5A–B). The growth, colony formation, migration and invasion of SK-OV-3 cell were detected using CCK-8, colony formation, wound healing and Transwell invasion assay. As shown in Figure 5C–D, over-expression of ELAVL1 restored the proliferation and colony formation of SK-OV-3 cell that was suppressed by miR-139-3p. Consistently, the migration and invasion of SK-OV-3 cell that was inhibited by miR-139-3p was rescued by up-regulation of ELAVL1 (Figure 5E–F). These findings indicated that miR-139-3p inhibited ovarian cancer cell progression through inhibiting ELAVL1 expression.
ELAVL1 Is Negatively Correlated With miR-139-3p In Ovarian Cancer
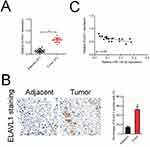
Finally, we analyzed the mRNA level of ELAVL1 in 21 cases of ovarian cancer and adjacent normal tissues using qRT-PCR. As shown in Figure 6A, ELAVL1 was over-expressed in the ovarian cancer tissue than that in the normal tissue. Immunohistochemical (IHC) staining assay proved that the expression of ELAVL1 was higher in ovarian cancer tissue than that in the normal tissue (Figure 6B). Furthermore, the Spearman correlation analysis suggested that the level of ELAVL1 was negatively correlated with miR-139-3p in ovarian cancer (Figure 6C).
Discussion
In the current study, we have demonstrated that miRNA-139-3p was down-expressed in ovarian cancer tissues and cells. Our investigation also proved that miRNA-139-3p suppressed the progression of ovarian cancer cell in vitro and in vivo. We also identified that ELAVL1 was a functional target gene of miRNA-139-3p and demonstrated that the level of miR-139-3p was negatively correlated with the level of ELAVL1 in ovarian cancer. Further investigation suggested that over-expression of ELAVL1 rescued the aggressiveness of ovarian cancer cell that was inhibited by miR-139-3p. Taken together, our study indicated that miRNA-139-3p inhibited the progression of ovarian cancer cell via inhibiting the expression of ELAVL1.
Aberrant expression of miRNA-139-3p has been reported in several cancers and miRNA-139-3p is widely considered to be a tumor suppressor in these cancers.13–16 MiR-139-3p was significantly down-regulated in colon cancer than that in noncancerous tissues and lower level of miR-139-3p was related with poor survival of patient with colorectal cancer.17 Furthermore, down-regulation of miR-139 was associated significantly with poor prognosis of patients with hepatocellular carcinoma (HCC) and the features of metastatic cancer, including microsatellite formation, reduced differentiation and absence of tumor encapsulation.18 Consistent with previous reports, we analyzed the expressions of miRNA-139-3p in ovarian cancer tissues and cell lines using qRT-PCR assay. Our data indicated that miR-139-3p was down-expressed in ovarian cancer cells and tissues.
To date, there are plenty of investigations demonstrating that miRNA-139-3p suppressed the metastatic behavior in several kinds of cancers. Up-regulation of miR-139-5p is associated with reduced metastatic activities in hepatocellular carcinoma (HCC) and breast cancer cell.19–21 In addition, in patients with invasive squamous cell carcinoma, lower level of miR-139-5p is associated with increased metastatic disease.7 To further reveal the function of miRNA-139-3p in the metastatic behavior of ovarian cancer, SK-OV-3 or OVCAR3 cell was transfected with miR-139-3p. Then, the growth, colony formation, migration and invasion abilities of ovarian cancer cell in vitro were analyzed. Consistent with previous reports, our data suggested that miRNA-139-3p served as a negative metastatic regulator in ovarian cancer.
MiRNAs imperfectly bind to the 3′-untranslated region (UTR) of target genes and induce translation inhibition of mRNAs degradation.22 Herein, we verified that ELAVL1 was the direct target of miRNA-139-3p in ovarian cancer. ELAVL1 (HuR), a nucleo-cytoplasmic shuttling protein, plays an important role in mRNA stability as well as cellular differentiation. The multiple functions of ELAVL1 in the development of cancer have been revealed.23–28 ELAVL1 is upregulated in human colorectal cancer and has been reported to promote colon cancer cell growth through regulating RNA in the cell cytoplasm.29 Moreover, ELAVL1 facilitates cancer stemness of lung cancer cell by modulating miR-873/CDK3 and miR-125a-3p/CDK3 axis.30 In breast cancer, miR-125a inhibits cell growth and promotion of apoptosis of cancer cell via targeting ELAVL1. Importantly, the repression of cell proliferation and migration engendered by miR-125a was partly rescued by ELAVL1 re-expression.31 In the present study, we found that miRNA-139-3p inhibited the expression of ELAVL1 in ovarian cancer cell and the level of miRNA-139-3p was negatively related with the level of ELAVL1 in ovarian carcinoma tissues. Our findings also indicated that miRNA-139-3p suppressed the aggressiveness of ovarian carcinoma through directly regulating ELAVL1.
Conclusion
In brief, our study demonstrated that miRNA-139-3p was markedly down-regulated in ovarian cancer and transfection of miR-139-3p inhibited the growth as well as metastasis of ovarian cancer cell through inhibiting the expression of ELAVL1.
Acknowledgments
This study was supported by the National Science Foundation of China (30901987); the Promotive research fund for excellent young and middle-aged scientisits of Shandong Province (BS2010YY034) and the Foundation of Mitochondrial Mechanism of SKOV3 apoptosis induced by resveratrol in Ovarian Cancer cells of Ji'nan Health Planning Commission (200705).
Disclosure
The authors declare that they have no competing interests in this work.
References
1. Mak CS, Yung MM, Hui LM, et al. MicroRNA-141 enhances anoikis resistance in metastatic progression of ovarian cancer through targeting KLF12/Sp1/survivin axis. Mol Cancer. 2017;16(1):11. doi:10.1186/s12943-017-0582-2
2. Zhang S, Zhang JY, Lu LJ, Wang CH, Wang LH. MiR-630 promotes epithelial ovarian cancer proliferation and invasion via targeting KLF6. Eur Rev Med Pharmacol Sci. 2017;21(20):4542–4547.
3. Chen MW, Yang ST, Chien MH, et al. The STAT3-miRNA-92-Wnt signaling pathway regulates spheroid formation and malignant progression in ovarian cancer. Cancer Res. 2017;77(8):1955–1967. doi:10.1158/0008-5472.CAN-16-1115
4. Li HY, Liang JL, Kuo YL, et al. miR-105/93-3p promotes chemoresistance and circulating miR-105/93-3p acts as a diagnostic biomarker for triple negative breast cancer. Breast Cancer Res. 2017;19(1):133. doi:10.1186/s13058-017-0918-2
5. Yu Y, Yin W, Yu ZH, et al. miR-190 enhances endocrine therapy sensitivity by regulating SOX9 expression in breast cancer. J Exp Clin Cancer Res. 2019;38(1):22. doi:10.1186/s13046-019-1039-9
6. Hui W, Ma X, Zan Y, et al. MicroRNA-1292-5p inhibits cell growth, migration and invasion of gastric carcinoma by targeting DEK. Am J Cancer Res. 2018;8(7):1228–1238.
7. Wang K, Jin J, Ma T, Zhai H. MiR-139-5p inhibits the tumorigenesis and progression of oral squamous carcinoma cells by targeting HOXA9. J Cell Mol Med. 2017;21(12):3730–3740. doi:10.1111/jcmm.13282
8. Chen H, Xu H, Meng YG, et al. miR-139-5p regulates proliferation, apoptosis, and cell cycle of uterine leiomyoma cells by targeting TPD52. Onco Targets Ther. 2016;9:6151–6160.
9. Miyoshi J, Toden S, Yoshida K, et al. MiR-139-5p as a novel serum biomarker for recurrence and metastasis in colorectal cancer. Sci Rep. 2017;7:43393.
10. Liu J, Li C, Jiang Y, et al. Tumor-suppressor role of miR-139-5p in endometrial cancer. Cancer Cell Int. 2018;18:51. doi:10.1186/s12935-018-0545-8
11. Zhang HD, Sun DW, Mao L, et al. MiR-139-5p inhibits the biological function of breast cancer cells by targeting Notch1 and mediates chemosensitivity to docetaxel. Biochem Biophys Res Commun. 2015;465(4):702–713. doi:10.1016/j.bbrc.2015.08.053
12. Livak KJ, Schmittgen TD. Analysis of relative gene expression data using real-time quantitative PCR and the 2(-Delta Delta C(T)) method. Methods. 2001;25(4):402–408. doi:10.1006/meth.2001.1262
13. Chen J, Yu Y, Chen X, et al. MiR-139-5p is associated with poor prognosis and regulates glycolysis by repressing PKM2 in gallbladder carcinoma. Cell Prolif. 2018;51(6):e12510. doi:10.1111/cpr.12510
14. Liu H, Yin Y, Hu Y, et al. miR-139-5p sensitizes colorectal cancer cells to 5-fluorouracil by targeting NOTCH-1. Pathol Res Pract. 2016;212(7):643–649. doi:10.1016/j.prp.2016.04.011
15. Song M, Yin Y, Zhang J, et al. MiR-139-5p inhibits migration and invasion of colorectal cancer by downregulating AMFR and NOTCH1. Protein Cell. 2014;5(11):851–861. doi:10.1007/s13238-014-0093-5
16. Zhang HD, Jiang LH, Sun DW, Li J, Tang JH. MiR-139-5p: promising biomarker for cancer. Tumour Biol. 2015;36(3):1355–1365. doi:10.1007/s13277-015-3199-3
17. Liu X, Duan B, Dong Y, et al. MicroRNA-139-3p indicates a poor prognosis of colon cancer. Int J Clin Exp Pathol. 2014;7(11):8046–8052.
18. Wong CC, Wong CM, Tung EK, et al. The microRNA miR-139 suppresses metastasis and progression of hepatocellular carcinoma by down-regulating Rho-kinase 2. Gastroenterology. 2011;140(1):322–331. doi:10.1053/j.gastro.2010.10.006
19. Qiu G, Lin Y, Zhang H, Wu D. miR-139-5p inhibits epithelial-mesenchymal transition, migration and invasion of hepatocellular carcinoma cells by targeting ZEB1 and ZEB2. Biochem Biophys Res Commun. 2015;463(3):315–321. doi:10.1016/j.bbrc.2015.05.062
20. Krishnan K, Steptoe AL, Martin HC, et al. miR-139-5p is a regulator of metastatic pathways in breast cancer. RNA. 2013;19(12):1767–1780. doi:10.1261/rna.042143.113
21. Dai H, Gallagher D, Schmitt S, et al. Role of miR-139 as a surrogate marker for tumor aggression in breast cancer. Hum Pathol. 2017;61:68–77. doi:10.1016/j.humpath.2016.11.001
22. Wang T, Hou J, Li Z, et al. miR-15a-3p and miR-16-1-3p negatively regulate twist1 to repress gastric cancer cell invasion and metastasis. Int J Biol Sci. 2017;13(1):122–134. doi:10.7150/ijbs.14770
23. Yuan Z, Sanders AJ, Ye L, Jiang WG. HuR, a key post-transcriptional regulator, and its implication in progression of breast cancer. Histol Histopathol. 2010;25(10):1331–1340. doi:10.14670/HH-25.1331
24. Kakuguchi W, Kitamura T, Kuroshima T, et al. HuR knockdown changes the oncogenic potential of oral cancer cells. Mol Cancer Res. 2010;8(4):520–528. doi:10.1158/1541-7786.MCR-09-0367
25. Allegri L, Mio C, Russo D, Filetti S, Baldan F. Effects of HuR downregulation on anaplastic thyroid cancer cells. Oncol Lett. 2018;15(1):575–579. doi:10.3892/ol.2017.7289
26. Brody JR, Dixon DA. Complex HuR function in pancreatic cancer cells. Wiley Interdiscip Rev RNA. 2018;9(3):e1469. doi:10.1002/wrna.1469
27. Liu Y, Chen X, Cheng R, et al. The Jun/miR-22/HuR regulatory axis contributes to tumourigenesis in colorectal cancer. Mol Cancer. 2018;17(1):11. doi:10.1186/s12943-017-0751-3
28. Sommer S, Cui Y, Brewer G, Fuqua SA. The c-Yes 3ʹ-UTR contains adenine/uridine-rich elements that bind AUF1 and HuR involved in mRNA decay in breast cancer cells. J Steroid Biochem Mol Biol. 2005;97(3):219–229. doi:10.1016/j.jsbmb.2005.09.002
29. Al-Haidari A, Algaber A, Madhi R, Syk I, Thorlacius H. MiR-155-5p controls colon cancer cell migration via post-transcriptional regulation of Human Antigen R (HuR). Cancer Lett. 2018;421:145–151. doi:10.1016/j.canlet.2018.02.026
30. Zhang Y, Yang L, Ling C, Heng W. HuR facilitates cancer stemness of lung cancer cells via regulating miR-873/CDK3 and miR-125a-3p/CDK3 axis. Biotechnol Lett. 2018;40(4):623–631. doi:10.1007/s10529-018-2512-9
31. Guo X, Wu Y, Hartley RS. MicroRNA-125a represses cell growth by targeting HuR in breast cancer. RNA Biol. 2009;6(5):575–583. doi:10.4161/rna.6.5.10079
 © 2019 The Author(s). This work is published and licensed by Dove Medical Press Limited. The full terms of this license are available at https://www.dovepress.com/terms.php and incorporate the Creative Commons Attribution - Non Commercial (unported, v3.0) License.
By accessing the work you hereby accept the Terms. Non-commercial uses of the work are permitted without any further permission from Dove Medical Press Limited, provided the work is properly attributed. For permission for commercial use of this work, please see paragraphs 4.2 and 5 of our Terms.
© 2019 The Author(s). This work is published and licensed by Dove Medical Press Limited. The full terms of this license are available at https://www.dovepress.com/terms.php and incorporate the Creative Commons Attribution - Non Commercial (unported, v3.0) License.
By accessing the work you hereby accept the Terms. Non-commercial uses of the work are permitted without any further permission from Dove Medical Press Limited, provided the work is properly attributed. For permission for commercial use of this work, please see paragraphs 4.2 and 5 of our Terms.