Back to Journals » Cancer Management and Research » Volume 12
MicroRNA-1276 Promotes Colon Cancer Cell Proliferation by Negatively Regulating LACTB
Authors Wang C, Shi Z, Hong Z, Pan J , Chen Z , Qiu C , Zhuang H, Zheng X
Received 24 August 2020
Accepted for publication 12 November 2020
Published 26 November 2020 Volume 2020:12 Pages 12185—12195
DOI https://doi.org/10.2147/CMAR.S278566
Checked for plagiarism Yes
Review by Single anonymous peer review
Peer reviewer comments 2
Editor who approved publication: Dr Eileen O'Reilly
Chunxiao Wang,* Zesheng Shi,* Zhongshi Hong, Jianpeng Pan, Zhichuan Chen, Chengzhi Qiu, Haibin Zhuang, Xuecong Zheng
Department of General Surgery, The Second Affiliated Hospital of Fujian Medical University, Quanzhou, People’s Republic of China
*These authors contributed equally to this work
Correspondence: Zhongshi Hong Department of General Surgery
The Second Affiliated Hospital of Fujian Medical University, No. 34 Zhongshan North Road, Licheng District, Quanzhou 362000 Fujian Province, People’s Republic of China
Tel +86 13859776152
Email [email protected]
Purpose: LACTB, regulated by a variety of microRNAs (miRNAs), is proven to be a tumor suppressor. However, there are few reports that LACTB in colon cancer cells is regulated by miRNA. Therefore, the aim of this study was to explore the miRNAs that regulate LACTB in colon cancer.
Patients and Methods: Data from TCGA were analyzed in starBase and GEPIA2, and Western blot and quantitative PCR (qPCR) were used to detect the expression of LACTB in colon cancer cell lines. MiRNAs targeting LACTB were predicted by MicroT-CDS, starBase, miRDB, mirDIP, and DIANA. The relationship between LACTB and miRNA was explored by dual-luciferase assay. MTT, propidium iodide (PI), Western blot, Annexin V-FITC/PI Kit, qPCR and transwell assay were used to detect the changes in cell proliferation, cell cycle, autophagy, apoptosis, epithelial-to-mesenchymal transition (EMT), cell migration, and invasiveness in colon cancer cells that overexpressed miR-1276 and/or LACTB.
Results: The results showed that the LACTB mRNA level was lower and the miR-1276 level was higher in colon cancer than in normal tissue. MiR-1276 inhibited the expression of LACTB. Furthermore, overexpression of miR-1276 in colon cancer cells increased proliferation, migration, invasiveness and EMT, and decreased autophagy and apoptosis. Supplementing LACTB suppressed these effects of miR-1276.
Conclusion: In conclusion, miR-1276, which may be a potential therapy for colon cancer, inhibits cell growth and promotes apoptosis by targeting LACTB in colon cancer cells.
Keywords: microRNA, colonic neoplasms, cell proliferation, LACTB, miR-1276
Introduction
Colon cancer is one of the deadliest, most common and increasingly prevalent malignancies, and it currently has the second-highest rate of cancer-related deaths worldwide.1–3 Despite the development of methods for early diagnosis and molecular therapeutic targets for colon cancer, the mortality rate among colon cancer patients remains high, making the development of more sensitive and specific diagnostic molecules, biomarkers, and therapeutic targets extremely urgent.1,4,5
Serine beta-lactamase-like protein (LACTB) is a mitochondrial protein that is most commonly expressed in the skeletal muscles, heart, and liver in mammals.6–8 It has been reported that LACTB is likely to regulate oxidative phosphorylation and mitochondrial lipid metabolism, thereby influencing tumor progression by influencing mitochondrial membrane tissue and microencapsulation.1,9 Studies have shown that LACTB has a strong inhibitory effect on a variety of tumors, including breast cancer, glioma, and colorectal cancer.1 LACTB can inhibit the epithelial-to-mesenchymal transition (EMT) and cell proliferation of breast cancer and colorectal cancer.1 In addition, studies have revealed that LACTB regulates the stability of p53 tumor suppressor to inhibit colorectal cancer invasion and migration.1,4,10–12
MicroRNA (miRNA), a type of endogenous non-coding RNA of approximately 22 nucleotides, has cell-specific or tissue-specific expression.13 It has the ability to regulate the stability and translation of messenger RNA (mRNA).13 It has been reported that miRNAs are involved in many biological processes, including migration, invasion, EMT, and metastasis.13 Among them, miRNA-1276 (miR-1276) is involved in many tumor processes by regulating certain genes, including gastric cancer, lung cancer, laryngeal cancer, and hepatocellular carcinoma.13–16 Moreover, many studies have found that LACTB is regulated by a variety of miRNAs, including miR-125b-5p, miR-351-5p, and miR-374a.12,17,18 However, there are few reports about miR-1276 regulating the expression of LACTB in colon cancer. Therefore, this study aims to explore the regulation of miR-1276 on LACTB expression and its influence on colon cancer cells.
Patients and Methods
Bioinformatics Analysis
RNA-seq data from patients with colon adenocarcinoma (COAD) from The Cancer Genome Atlas (TCGA) were analyzed in starBase (http://starbase.sysu.edu.cn/) and GEPIA2 (http://gepia2.cancer-pku.cn/#analysis). An in-depth analysis was performed using GraphPad Prism 8.2.1 to compare the differences of LACTB levels in tumor of different stages. Patients with COAD were divided into two groups based on the average value of miR-1276 level or the expression level (FPKM) of LACTB at 4.0 to compare perform Kaplan-Meier (KM) curve analysis in the starBase database or The Human Protein Atlas database, respectively. MiRNA data targeting LACTB which were predicted in MicroT-CDS, starBase, miRDB, DIANA, and mirDIP were screened using UpSetR package in R version 3.5.1.
Cell Lines
Cell lines, HCT-8, HCT-116 and Caco-2, purchased from American Type Culture Collection (ATCC, Manassas, VA 20110 USA), were cultured in RPMI-1640 medium (ATCC, Catalog No. 30-2001) supplemented with 10% horse serum (GIBCO, Catalog No. 26050070, Shanghai, China), McCoy’s 5a medium (ATCC, Catalog No. 30–2007) supplemented with 10% fetal bovine serum (FBS) (GIBCO, Catalog No. 10091-148, Shanghai, China), and Eagle’s Minimum Essential Medium (ATCC, Catalog No. 30-2003) supplemented with 20% FBS, respectively. Cell lines, SW480, SW620, and NCM460, obtained from Xiamen Immocell Biotechnology Co., Ltd (Xiamen, Fujian, China), were cultured in Dulbecco’s modified eagle medium (DMEM) (GIBCO, Catalog No. 11965-092, Shanghai, China) supplemented with 10% FBS. All cell lines were maintained in a humidified incubator at 37°C with 5% CO2.
Quantitative PCR (qPCR)
TRIzol reagent (Invitrogen, Catalog No. 15596026, Shanghai, China) was used to extract total RNA, which was then reverse transcribed into cDNA by Hiscript Reverse Transcriptase (VAZYME, Catalog No. R101-01/02, Nanjing, Jiangsu, China). Subsequently, according to the manufacturer’s instructions, qPCR was performed with AceQ qPCR SYBR Green Master Mix (VAZYME, Catalog No. Q111-02, Nanjing, Jiangsu, China) in an iQ5 Real-Time PCR Detection System (Bio-Rad Laboratories, Hercules, CA, USA) using the obtained cDNA to determine relative RNA expression levels. The primers for qPCR are shown in Table 1. Finally, the target gene expression was quantified using the 2−ΔΔCt method.
 |
Table 1 Primers for qPCR and Reverse Transcription |
Western Blot
Ice-cold RIPA buffer (Beyotime Biotechnology, Catalog No. P0013C, Shanghai, China) and the BCA protein assay kit (Beyotime Biotechnology, Catalog No. P0012S, Shanghai, China) were used to extract and quantify proteins, respectively. Afterward, Western blotting was performed as previously described.19 The primary antibodies used were LACTB Antibody (Proteintech, Catalog No. 66785-1-Ig, 1:3000, Wuhan, Hubei, China), LC3 Antibody (Proteintech, Catalog No. 14600-1-AP, 1:1000, Wuhan, Hubei, China), P62 Antibody (Proteintech, Catalog No. 18420-1-AP, 1:1000, Wuhan, Hubei, China) and GAPDH Antibody (Proteintech, Catalog No. 60004-1-Ig, 1:20,000, Wuhan, China). The secondary antibodies were HRP-conjugated Affinipure Goat Anti-Mouse IgG (Proteintech, Catalog No. SA00001-1, 1:2000, Wuhan, Hubei, China) and HRP-conjugated Affinipure Goat Anti-Rabbit IgG (Proteintech, Catalog No. SA00001-2, 1:2000, Wuhan, Hubei, China).
Construction of Plasmids
Wild type or mutant of the LACTB 3ʹUntranslated Regions (3ʹUTR) were cloned into pmirGLO vectors, which were acquired from XIAMEN Anti-HeLa Biological Technology Trade Co. Ltd (Xiamen, Fujian, China) and named LACTB 3ʹUTR WT and LACTB 3ʹUTR MUT, respectively. The LACTB 3ʹUTR mutation site is shown in red letters in Figure 1B. The corresponding primers were designed using DNAMAN 10.0 and are shown in Table 2.
 |
Table 2 Primers for Plasmid Construction |
LACTB and miR-1276 were cloned into pCDH vectors, purchased from XIAMEN Anti-HeLa Biological Technology Trade Co. Ltd (Xiamen, Fujian, China), to prepare the LACTB and miR-1276 expression plasmids, respectively. The corresponding primer sequences were designed using DNAMAN 10.0 (Table 2).
Dual-Luciferase Reporter Assay
HCT-8 and HCT-116 cells were incubated overnight in 24-well plates at a density of 4 × 104 cells per well. Plasmid LACTB 3ʹUTR WT or LACTB 3ʹUTR MUT was co-transfected into cells with the negative control of miR-1276 plasmid or miR-1276 plasmid using the Xfect Transfection Reagent (VAZYME, Catalog No. T101-01, Nanjing, Jiangsu, China) according to the manufacturer’s instructions. After 48 h, a dual-Luciferase Reporter Assay (Promega, Catalog No. E1910, Beijing, China) was performed following the manufacturer’s instructions.
MTT Assay
HCT-8 and HCT-116 cells, co-transfected with MiR-1276 plasmid and LACTB plasmid or their negative control for 24 h, were cultured in 96-well plates at a density of 1×104 cells per well. After 24 hours, the MTT assay was performed as previously described.20 Absorbance was measured at OD450 on a microplate reader (Thermo, Shanghai, China).
Apoptosis Assay
HCT-8 and HCT-116 cells, transfected with plasmids as above described, were cultured in 6-well plates at a density of 5×105 cells per well. After 48 h, cells were stained using Annexin V-FITC/propidium iodide (PI) Apoptosis Detection Kit (VAZYME, Catalog No. A211-01, Nanjing, Jiangsu, China) as previously described.21 Flow cytometry (ACEA Biosciences, Inc., San Diego, CA, USA) was then used for detection.
Cell Cycle Assay
HCT-8 and HCT-116 cells were evenly plated in 6-well plates. MiR-1276 plasmid and its negative control were transfected into cells with a plasmid expressing LACTB or the negative control of LACTB using Xfect Transfection Reagent. After 24 h, cell cycle assay was performed using PI and a flow cytometer (ACEA Biosciences, Inc., San Diego, CA, USA) as previously described.22
Transwell Assay
After HCT-8 and HCT-116 cells were transfected with plasmids as above described, transwell plates with and without Matrigel were used for migration and invasion, respectively, by transwell assay as previously described.19,23 After 24 h, migrated or invasive cells were evaluated through 0.5% crystal violet staining, photographed using a light microscope (MOTIC, Hongkong, China), and counted.
Statistical Analysis
SPSS 22.0 (IBM SPSS, Armonk, New York, USA) was used to analyze all statistics, Student’s t-test (unpaired) was used to analyze the difference between the two groups, and analysis of variance (ANOVA) was used to analyze the multiple groups, with p <0.05 defined as a significant difference. GraphPad Prism 8.2.1 was used to draw a graph. Data are expressed as mean ± standard deviation (SD).
Results
LACTB Level Was Lower in Colon Cancer Than That in Normal Tissue
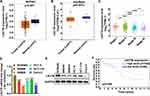
The mRNA level of LACTB in colon cancer tissues and normal tissues from TCGA and starBase was analyzed, finding that the mRNA level of LACTB in colon cancer tissues is lower than that in normal tissues, and the mRNA level of LACTB is associated with the TNM stage of colon cancer (Figure 2A–C). Next, the mRNA and protein levels of LACTB were measured in colon cancer cell lines (SW480, SW620, HCT-8, HCT-116, and Caco-2) and a normal colon epithelial cell line (NCM460). The result showed that the mRNA and protein levels of LACTB are lower in SW480, SW620, HCT-8, HCT-116, and Caco-2 than in NCM460 (Figure 2D and E), which is consistent with the above results. The KM curve analysis from The Human Protein Atlas (THPA) showed that compared with patients with high-level LACTB, patients with low-level LACTB have worse overall survival (Figure 2F). These findings suggested that in colon cancer, low levels of LACTB are likely to have an impact on tumor progression and indicate poor prognosis.
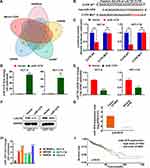
MiR-1276 Targets LACTB
MiRNAs targeting LACTB were screened from MicroT-CDS, starBase, miRDB, mirDIP, and DIANA databases, and showed miR-1276 targeting LACTB in all five databases (Figure 1A). By comparing the miR-1276 sequence with LACTB 3ʹ UTR, we found the site where miR-1276 may targets (Figure 1B). Then, we constructed the mutated LACTB 3ʹUTR by producing mutation at the site where LACTB 3ʹUTR targets to further verify this site and the regulatory effect of miR-1276 on LACTB by dual-luciferase reporter assay. The dual-luciferase reporter assay showed that miR-1276 inhibited the wild-type LACTB 3ʹUTR, but not the LACTB 3ʹUTR mutant (Figure 1C). Transfection of the plasmid expressing miR-1276 in HCT-8 and HCT-116 cells increased the level of miR-1276, while decreasing the mRNA and protein levels of LACTB (Figure 1D–F). By analyzing the data from TCGA, it was found that the level of miR-1276 in colon cancer tissues was higher than that in normal tissues (Figure 1G). Moreover, the miR-1276 level in SW480, SW620, HCT-8, HCT-116, and Caco-2 was higher than that in NCM460 (Figure 1H). Through the KM curve analysis, it was found that the level of miR-1276 had no effect on the survival time of colon cancer patients (Figure 1I).
MiR-1276 Negatively Regulates LACTB to Promote the Proliferation
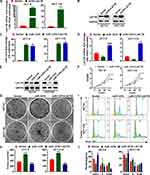
To investigate the effect of miR-1276 negatively regulating LACTB on cell biological processes, a plasmid expressing LACTB was constructed. As shown in Figure 3A and B, the plasmid effectively increased the mRNA and protein levels of LACTB in HCT-8 and HCT-116 cells (Figure 3A and B). Subsequently, the cells were treated with miR-1276 and plasmids individually or together, and it was found by qPCR and Western blot that miR-1276 reduced the mRNA and protein levels of LACTB, whereas after supplementing LACTB, the expression level of LACTB increased (Figure 3C–E). Furthermore, increasing the level of miR-1276 accelerated the proliferation of cells, increased the number of cell colonies, and promoted the conversion of the G0/G1 phase to the S phase of the cell cycle, while the supplementation of LACTB retarded down these effects of miR-1276 on cells (Figure 3F–J). These findings revealed that miR-1276 targets LACTB to improve proliferation.
MiR-1276 Suppresses the Apoptosis by Negatively Regulating LACTB
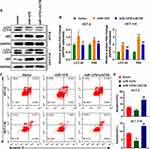
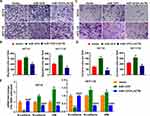
After overexpression of miR-1276, the ratio of the protein levels of LC3I to LC3II in the cells was increased, and simultaneously, the protein level of p62 was increased (Figure 4A and B). After supplementation of LACTB, the ratio of the protein levels of LC3I to LC3II and the protein level of p62 were both decreased, suggesting that miR-1276 inhibits autophagy by reducing the level of LACTB (Figure 4A and B).
Moreover, after miR-1276 treatment, the apoptosis level of HCT-8 and HCT-116 cells was decreased, while apoptosis level was elevated after LACTB supplementation (Figure 4C and D). These results showed that miR-1276 suppresses apoptosis by negatively regulating LACTB. In short, miR-1276 hindered autophagy by negatively regulating LACTB, thereby inhibiting cell apoptosis.
Replenishment of LACTB Disrupts miR-1276-Induced Migration and Invasion
Afterward, migrated and invasive cells were also detected using a transwell assay. The results showed that miR-1276 enhanced cell migration ability and invasiveness, while the amount of migrating and invasive cells decreased after overexpression of LACTB (Figure 5A–D).
Furthermore, increasing miR-1276 reduced the mRNA level of E-cadherin and elevated the mRNA levels of N-cadherin and Vimentin (Figure 5E). After supplementing LACTB, the mRNA level of E-cadherin was increased, and the mRNA levels of N-cadherin and Vimentin were decreased (Figure 5E), indicating that miR-1276 facilitates EMT by lowering LACTB levels.
Discussion
Colon cancer is an aggressive tumor with high morbidity and mortality.1 The distant metastasis of the tumor seriously impairs the survival of colon cancer patients, which suggests that the molecular mechanism of colon cancer invasion needs to be clarified urgently.1 Many studies have proved that LACTB can inhibit tumor development.6 Moreover, studies have reported that LACTB is significantly down-regulated in colorectal cancer, and the low expression of LACTB correlates with the poor overall survival of colorectal cancer patients, which is consistent with this study’s findings.9 In addition, low levels of LACTB are associated with colorectal cancer metastasis and advanced clinical stages.9 LACTB has also been identified as an independent prognostic factor for colorectal cancer.9 Overexpression of LACTB in vitro promotes autophagy of colon cancer cells and inhibits the proliferation, migration, invasion, and EMT of colon cancer cells.1,9 Consistent with this, we observed that miR-1276 suppresses autophagy and accelerates EMT by lowering LACTB levels. In vivo experiments confirmed that overexpression of LACTB can inhibit the growth and metastasis of colorectal cancer.9 These findings indicate that LACTB can be another strategy for the treatment of colon cancer.
Previous studies have found multiple ways in which LACTB inhibits tumor progression.1 LACTB inhibits the progression of colorectal cancer by attenuating the MDM2-mediated p53 ubiquitination.1,7,24 LACTB promotes autophagy of colorectal cancer cells by regulating the activity of PIK3R3 and by inhibiting EMT and proliferation.1 LACTB inhibits tumors by increasing mitochondrial lipid metabolism.11 MiRNAs are a class of conservative non-coding RNAs, which promote mRNA degradation or inhibit the translation of target genes by binding to the 3ʹUTR region of target genes, thereby inhibiting the expression of target proteins.12,25 Accumulating evidence has shown that LACTB is also regulated by miRNA. LACTB is regulated by miR-125b-5p to reduce the secretion of MCP-1 in macrophages.17 MiR-351-5p directly targets LACTB to mediate skeletal muscle production and miR-374a promotes breast cancer metastasis and progression by down-regulating LACTB.12,18
In addition, more and more studies have found that a large number of miRNAs are involved in cancer development and progression.12,26,27 MiR-1276 is also involved in the regulation of cancer cells, for example, MiR-1276 is a target of NF-κB, which can help tumor necrosis factor α promote adriamycin-induced apoptosis of cancer cells by positively regulating CASP9 and miR-1276 regulates the expression of CTNNB1 through the regulation of LncRNA HCG11, thus affecting the proliferation and migration of gastric cancer cells.14,28 It was found that miR-1276 is highly expressed in colon cancer cells, and promotes cell proliferation, migration, and invasion, while inhibiting cell apoptosis. In this study, we found that LACTB is also negatively regulated by miR-1276 in colon cancer cells.
In the current study, the KM curve analysis from THPA shows that the overall survival of patients with low-level LACTB is worse than that of patients with high-level LACTB, while the KM curve analysis grouped by the median value of miR-1276 showed that the overall survival of patients was not significantly affected by the miR-1276 level, which may be related to the fact that not only LACTB is regulated by miR-1276.
Conclusions
In conclusion, this study found that miR-1276 was highly expressed in colon cancer, and promoted colon cancer cell proliferation by negatively regulating LACTB. Moreover, LACTB has been confirmed to be an anticancer gene, and supplementing LACTB can reduce the cell proliferation, migration, and invasion induced by miR-1276. These findings provided new insights into the regulation of colon cancer.
Data Sharing Statement
The analyzed data sets generated during the present study are available from the corresponding author upon request.
Acknowledgments
Thank you for the financial support from the Science and Technology Project of Quanzhou (grant number 2018Z127), the Educational Research Project for Young and Middle-aged Teachers of Fujian Educational Committee (grant number JAT190216) and Startup Fund for scientific research of Fujian Medical University (grant number 2019QH1119).
Disclosure
All authors declare that there is no conflict of interest in this study.
References
1. Xu W, Yu M, Qin J, Luo Y, Zhong M. LACTB regulates PIK3R3 to promote autophagy and inhibit EMT and proliferation through the PI3K/AKT/mTOR signaling pathway in colorectal cancer. Cancer Manag Res. 2020;12:5181–5200. doi:10.2147/CMAR.S250661
2. Siegel RL, Miller KD, Jemal A. Cancer statistics, 2019. CA Cancer J Clin. 2019;69(1):7–34. doi:10.3322/caac.21551
3. Wingo PA, Cardinez CJ, Landis SH, et al. Long-term trends in cancer mortality in the United States, 1930–1998. Cancer. 2003;97(12 Suppl):3133–3275. doi:10.1002/cncr.11380
4. Kahi CJ, Boland CR, Dominitz JA, et al. Colonoscopy surveillance after colorectal cancer resection: recommendations of the US Multi-Society Task Force on Colorectal Cancer. Gastroenterology. 2016;150(3):758–768.e711. doi:10.1053/j.gastro.2016.01.001
5. Brody H. Colorectal cancer. Nature. 2015;521(7551):S1. doi:10.1038/521S1a
6. Keckesova Z, Donaher JL, De Cock J, et al. LACTB is a tumour suppressor that modulates lipid metabolism and cell state. Nature. 2017;543(7647):681–686. doi:10.1038/nature21408
7. Smith TS, Southan C, Ellington K, Campbell D, Tew DG, Debouck C. Identification, genomic organization, and mRNA expression of LACTB, encoding a serine beta-lactamase-like protein with an amino-terminal transmembrane domain. Genomics. 2001;78(1–2):12–14. doi:10.1006/geno.2001.6643
8. Mootha VK, Bunkenborg J, Olsen JV, et al. Integrated analysis of protein composition, tissue diversity, and gene regulation in mouse mitochondria. Cell. 2003;115(5):629–640. doi:10.1016/S0092-8674(03)00926-7
9. Zeng K, Chen X, Hu X, et al. LACTB, a novel epigenetic silenced tumor suppressor, inhibits colorectal cancer progression by attenuating MDM2-mediated p53 ubiquitination and degradation. Oncogene. 2018;37(41):5534–5551. doi:10.1038/s41388-018-0352-7
10. Xue C, He Y, Zhu W, et al. Low expression of LACTB promotes tumor progression and predicts poor prognosis in hepatocellular carcinoma. Am J Transl Res. 2018;10(12):4152–4162.
11. Cucchi D, Mauro C. LACTB-mediated tumour suppression by increased mitochondrial lipid metabolism. Cell Death Differ. 2017;24(7):1137–1139. doi:10.1038/cdd.2017.60
12. Zhang J, He Y, Yu Y, et al. Upregulation of miR-374a promotes tumor metastasis and progression by downregulating LACTB and predicts unfavorable prognosis in breast cancer. Cancer Med. 2018;7(7):3351–3362. doi:10.1002/cam4.1576
13. Zhan JW, Jiao DM, Wang Y, et al. Integrated microRNA and gene expression profiling reveals the crucial miRNAs in curcumin anti-lung cancer cell invasion. Thorac Cancer. 2017;8(5):461–470. doi:10.1111/1759-7714.12467
14. Zhang H, Huang H, Xu X, et al. LncRNA HCG11 promotes proliferation and migration in gastric cancer via targeting miR-1276/CTNNB1 and activating Wnt signaling pathway. Cancer Cell Int. 2019;19:350. doi:10.1186/s12935-019-1046-0
15. Lu E, Su J, Zhou Y, Zhang C, Wang Y. CCL20/CCR6 promotes cell proliferation and metastasis in laryngeal cancer by activating p38 pathway. Biomed Pharmacother. 2017;85:486–492. doi:10.1016/j.biopha.2016.11.055
16. Xiong DD, Dang YW, Lin P, et al. A circRNA-miRNA-mRNA network identification for exploring underlying pathogenesis and therapy strategy of hepatocellular carcinoma. J Transl Med. 2018;16(1):220.
17. Lu JB, Yao XX, Xiu JC, Hu YW. MicroRNA-125b-5p attenuates lipopolysaccharide-induced monocyte chemoattractant protein-1 production by targeting inhibiting LACTB in THP-1 macrophages. Arch Biochem Biophys. 2016;590:64–71. doi:10.1016/j.abb.2015.11.007
18. Du J, Zhang P, Zhao X, et al. MicroRNA-351-5p mediates skeletal myogenesis by directly targeting lactamase-β and is regulated by lnc-mg. FASEB J. 2019;33(2):1911–1926. doi:10.1096/fj.201701394RRR
19. Li Y, Wang Y, Fan H, Zhang Z, Li N. miR-125b-5p inhibits breast cancer cell proliferation, migration and invasion by targeting KIAA1522. Biochem Biophys Res Commun. 2018;504(1):277–282. doi:10.1016/j.bbrc.2018.08.172
20. Kumar P, Nagarajan A, Uchil PD. Analysis of cell viability by the MTT assay. Cold Spring Harb Protoc. 2018;2018(6).
21. Li T, Chen Y, Zhang J, Liu S. LncRNA TUG1 promotes cells proliferation and inhibits cells apoptosis through regulating AURKA in epithelial ovarian cancer cells. Medicine (Baltimore). 2018;97(36):e12131. doi:10.1097/MD.0000000000012131
22. Wu YS, Quan Y, Zhang DX, Liu DW, Zhang XZ. Synergistic inhibition of breast cancer cell growth by an epigenome-targeting drug and a tyrosine kinase inhibitor. Biol Pharm Bull. 2017;40(10):1747–1753. doi:10.1248/bpb.b17-00360
23. He JH, Li YG, Han ZP, et al. The CircRNA-ACAP2/Hsa-miR-21-5p/Tiam1 regulatory feedback circuit affects the proliferation, migration, and invasion of colon cancer SW480 cells. Cell Physiol Biochem. 2018;49(4):1539–1550. doi:10.1159/000493457
24. Torrano V, Carracedo A. Quiescence-like metabolism to push cancer out of the race. Cell Metab. 2017;25(5):997–999. doi:10.1016/j.cmet.2017.04.027
25. Lewis BP, Burge CB, Bartel DP. Conserved seed pairing, often flanked by adenosines, indicates that thousands of human genes are microRNA targets. Cell. 2005;120(1):15–20.
26. Farazi TA, Hoell JI, Morozov P, Tuschl T. MicroRNAs in human cancer. Adv Exp Med Biol. 2013;774:1–20.
27. Bertoli G, Cava C, Castiglioni I. MicroRNAs: new biomarkers for diagnosis, prognosis, therapy prediction and therapeutic tools for breast cancer. Theranostics. 2015;5(10):1122–1143. doi:10.7150/thno.11543
28. Zhou F, Li Y, Huang Y, et al. Upregulation of CASP9 through NF-κB and its target MiR-1276 contributed to TNFα-promoted apoptosis of cancer cells induced by doxorubicin. Int J Mol Sci. 2020;21(7).
 © 2020 The Author(s). This work is published and licensed by Dove Medical Press Limited. The full terms of this license are available at https://www.dovepress.com/terms.php and incorporate the Creative Commons Attribution - Non Commercial (unported, v3.0) License.
By accessing the work you hereby accept the Terms. Non-commercial uses of the work are permitted without any further permission from Dove Medical Press Limited, provided the work is properly attributed. For permission for commercial use of this work, please see paragraphs 4.2 and 5 of our Terms.
© 2020 The Author(s). This work is published and licensed by Dove Medical Press Limited. The full terms of this license are available at https://www.dovepress.com/terms.php and incorporate the Creative Commons Attribution - Non Commercial (unported, v3.0) License.
By accessing the work you hereby accept the Terms. Non-commercial uses of the work are permitted without any further permission from Dove Medical Press Limited, provided the work is properly attributed. For permission for commercial use of this work, please see paragraphs 4.2 and 5 of our Terms.