Back to Journals » OncoTargets and Therapy » Volume 14
MicroRNA-1249 Targets G Protein Subunit Alpha 11 and Facilitates Gastric Cancer Cell Proliferation, Motility and Represses Cell Apoptosis
Authors Zhang H, Fu T, Zhang C
Received 17 July 2020
Accepted for publication 22 January 2021
Published 24 February 2021 Volume 2021:14 Pages 1249—1259
DOI https://doi.org/10.2147/OTT.S272599
Checked for plagiarism Yes
Review by Single anonymous peer review
Peer reviewer comments 3
Editor who approved publication: Dr Federico Perche
Hongzhu Zhang,1,* Tingting Fu,1,* Cuiping Zhang2
1Department of Gastroenterology, Jinan Jigang Hospital, Jinan, Shandong, 250101, People’s Republic of China; 2Department of Gastroenterology, The Affiliated Hospital of Qingdao University, Qingdao, Shandong, 266000, People’s Republic of China
*These authors contributed equally to this work
Correspondence: Cuiping Zhang
Department of Gastroenterology, The Affiliated Hospital of Qingdao University, 16 Jiangsu Road, Shinan District, Qingdao, Shandong, 266000, People’s Republic of China
Email [email protected]
Aim: The purpose of our study was to investigate the effects of miR-1249 in gastric cancer.
Methods: By analyzing the data obtained from TCGA database, the expression and prognosis of miR-1249 in gastric cancer patients were analyzed. Then, CCK8, colony forming and transwell assays were used to test cell proliferation and motility. The cell apoptosis was detected by flow cytometry. The Pearson correlation coefficient analyzed was applied to analyze the correlation between GNA11 and miR-1249. qRT-PCR and Western blotting assays were employed to detect the mRNA and protein levels.
Results: We discovered that miR-1249 was highly expressed and was associated with a worse prognosis in gastric cancer patients. Besides, miR-1249 was up-regulated in gastric cancer cell lines (AGS, MKN45 and SNU1). More interestingly, miR-1249 exerted facilitating impacts on gastric cancer cell proliferation and motility, whereas miR-1249 acted as a suppressing effect on gastric cancer apoptosis. G protein subunit alpha 11 (GNA11) was a target gene of miR-1249 and was negatively correlated with miR-1249. Furthermore, GNA11 was negatively regulated by miR-1249. Additionally, GNA11 was lowly expressed in gastric cancer tissues and cell lines, as well as low GNA11 expression, was related to poor overall survival results in gastric cancer patients. The promoting influences of miR-1249 over-expression on AGS cell proliferation and motility was rescued by GNA11 over-expression, which might be achieved by regulating PI3K/AKT/mTOR signalling pathway.
Conclusion: Above all, we concluded that miR-1249 was concerned with the progression of gastric cancer through regulating GNA11, suggesting that miR-1249 and GNA11 might serve as predictive biomarkers for gastric cancer therapy.
Keywords: gastric cancer, miR-1249 mimic/inhibitor, G protein subunit alpha 11, target, proliferation, apoptosis, PI3K/AKT/mTOR pathway
Introduction
Gastric cancer is the fourth most common cancer in the world. According to its overall morbidity and mortality, it ranks second among all malignant tumors and is the leading cause of cancer-related death around the world.1–3 In 2018, there were approximately 1,033,701 new patients and 782,685 new deaths occurred around the whole world.4 Its clinical characteristics are usually proved for high recurrence rate, occult attacks, missed diagnosis, etc.5 Early clinical examination of gastric cancer is less than 15%, and advanced gastric cancer accounts for 85%.6 Currently, the main treatments for this disease are surgery, chemotherapy, radiation therapy, targeted therapy and immune gene therapy.7 Although these treatments have made great progress, due to chemotherapy resistance, the survival rate of patients with advanced or relapse is low, and the 5-year survival rate is less than 30%.8–10 Therefore, early diagnosis of gastric cancer is a major challenge and there is an urgent need to find more early diagnostic markers and solve the problem of chemical resistance. In recent years, more and more reports have reported that many miRNAs are closely related to gastric cancer,11–13 but the specific molecular mechanism of their roles in gastric cancer cells is still under studying.
As a series of small non-coding RNAs, microRNAs (miRNAs), with 18–25 nt, act important influences in posttranscriptional genes by suppressing the translation of their target mRNAs.14 Growing evidence has verified that miRNAs are related to various pathological and physiological processes including cell proliferation, differentiation, migration, invasion, metabolism, apoptosis and so on.15 Previous studies have revealed that miRNAs can serve as agonist or antagonist in the progression of various cancers.16–18 Furthermore, studies also have affirmed that some miRNAs are associated with various biological processes in gastric cancer.19,20 Utilizing bioinformatics analysis, we identified that miR-1249 was an agonist miRNA affecting gastric cancer development. Dang et al21 demonstrated that miR-1249 exerted repressive impacts on colon adenocarcinoma cell proliferation and motility, which might be possibly achieved by regulating Four-Jointed Box Kinase 1 (FJX1). Furthermore, miR-1249 was reported to be involved in hepatocellular carcinoma cell invasion and migration through targeting PTCH1.22 These data aroused us lots of interest in probing the influences of miR-1249 on gastric cancer.
Studies have indicated that various genes expressions are partially regulated by miRNAs.23,24 Utilizing bioinformatics analysis, G protein subunit alpha 11 (GNA11) was identified as one of the target genes of miR-1249 in our paper. Belongs to the family of heterotrimeric guanine nucleotide-binding protein G, GNA11 encodes the subunit α11.25 In immunocompromised mice, the GNA11Q209L and GNA11R183C mutants were able to transform 3T3 cells and took shape into tumors.26 There is one study has shown that GNA11 can activate somatic mutations at high frequency in uveal melanoma.25 Based on bioinformatics analysis, GNA11 was identified as one of the molecular targets of Korean patients with gastrointestinal stromal tumors.27 However, the mechanism of GNA11 on gastric cancer has not been fully elucidated, and further researches are needed.
In the present study, we aimed to explore the impacts of miR-1249 and GNA11 on gastric cancer development. Besides, we focused on investigating the interaction between miR-1249 and GNA11 in gastric cancer cells. We discovered a novel regulatory axis composed of miR-1249/GNA11, which might be possibly associated with PI3K/AKT/mTOR pathway, offering novel therapeutic targets and potential biomarkers for gastric cancer diagnosis and treatment.
Methods
Analyses of the Expression of miR-1249 and GNA11 Utilizing TCGA Database
First, the common miRNAs were obtained from the intersection miRNAs that had meaningful survival and differential expression miRNAs in gastric cancer, which were gained from TCGA database (https://cancergenome.nih.gov/). The miR-1249 expression data were also acquired from TCGA database. Moreover, the overall survival of miR-1249 on gastric cancer patients was analyzed based on TCGA dataset.
Next, we used the Veen diagram to identify the potential target genes of miR-1249. The common genes were obtained from the interaction among survival differences genes in TCGA database (ven_sur_allRNA), the expression difference genes in TCGA database (ven_dif_mRNA), the most related genes to hsa-miR-1249 by utilizing the pearson correlation analysis (ven_cor_best) and the predicted target genes from the TargetScan software (target_gene). Furthermore, according to the data acquired from TCGA database, the expression level of GNA11 was explored. Then, the overall survival analysis of GNA11 was implemented for gastric cancer patients based on TCGA datasets. Pearson’s correlation analysis was executed to analyze the relationship between miR-1249 and GNA11.
Culture of Gastric Cancer Cell Lines
Gastric cancer cell lines AGS, MKN45, SNU1, and the normal human gastric cell line GES-1 were acquired from Cell Bank of Chinese Academy of Medical Sciences (Shanghai, China). Then, these cells were cultivated in DMEM supplemented with 10% fetal bovine serum (FBS), 100 mg streptomycin per mL and 100 units penicillin per Ml under normal conditions.
Cell Transfection
Synthetic miR-1249 mimic/inhibitor and their corresponding negative control (mimic NC/inhibitor NC), pcDNA3.1-GNA11, pcDNA3.1-, si-GNA11 (5ʹ- ATTGCAGGGGATTGCCTTGG-3ʹ) and si-control (5ʹ- CGAACUCACUGGUCUGACC-3ʹ) were acquired from Dharmacon (Boulder, CO). Then, they were transfected into gastric cancer cells utilizing Lipofectamine 3000 reagent (Invitrogen). After 48 hours, the efficiency of transfection was determined by qRT-PCR.
qRT-PCR
Total RNA was extracted using TRIzol (Invitrogen). By employing Mir-X ™ miRNA First Strand Synthesis Kit or PrimeScript RT Reagent Kit, reverse transcription was conducted. SYBR PrimeScriptTM miRNA RT-PCT Kit or SYBR Premix Ex Taq II were applied to detect miR-1249 or GNA11 expression in an AB7300 real-time PCR machine. U6 served as a reference for miR-1249 detection. GAPDH was utilized as an internal control for GNA11 detection. 2−ΔΔCt was used to analyze the levels of miR-1249 and GNA11. Table 1 displays the primers used for qPCR.
 |
Table 1 The Primers Used in qRT-PCR |
Western Blotting Assay
Total proteins were isolated by a lysis buffer (containing protease suppressor). The concentration of the proteins was tested by BCA method. Then, the protein lysates were separated by SDS-PAGE and transferred onto a PVDF membrane. After being blocked by 5% defatted milk, the membrane was incubated with primary antibodies overnight at 4°C. Subsequently, the membrane was incubated with peroxidase-conjugated secondary antibodies at room temperature for 1 hour. The signals were tested utilizing an ECL detection kit (GE Healthcare). The quantitative analysis was normalized to GAPDH.
Cell Counting Kit-8 (CCK8)
Cell proliferation was evaluated by CCK8 assay (Beyotime, Shanghai, China) following the protocol. In brief, 1 × 103 cells/well were seeded in a 96-well plate. After transfection, 10 µL CCK8 reagent was added to each well and further cultured -. Finally, the OD450 was measured by a microplate reader (Biotech).
Colony Formation Assay
The transfected cells (500 cells/well) were plated into a 6-well plate and incubated for 14 days until a colony formed. After fixed with methanol for 15 min, the cells were stained with crystal violet. Finally, the number of colonies was counted under a microscope.
Transwell Assay
The transwell assay (Corning, New York, USA) was executed to test the abilities of cell invasion and migration. Briefly, the invasion and migration of gastric cancer cells (2 × 105/well) were measured by transwell inserts pre-coated with or without Matrigel. Besides, the top side of the transwell chamber was filled with DMEM without serum. The low side was filled with a 10% FBS medium. Only the invasion assay needs to pre-coat Matrigel on the upper chamber. After being washed with PBS, the invasive and migratory cells were fixed with 4% paraformaldehyde for 30 minutes and stained with 0.1% crystal violet for 20 minutes. Finally, the cells were photographed by an inverted microscope (Olympus) and the cell number was counted in five random fields.
Analysis of Apoptotic Cells
After 48 h transfection, the number of apoptotic gastric cancer cells was assessed by flow cytometry. In short, the transfected gastric cancer cells were washed with PBS. Then, the cells were dyed with 5 µL Annexin V-FITC for 10 minutes and stained with 10 µL PI for 5 minutes at room temperature under normal conditions devoid of light. Finally, the results were analyzed by a Flow cytometer.
Luciferase Reporter Assay
After 48 h transfection, the gastric cancer cells (1×104/well) were seeded into 96-well plates. After 24 hr incubation, the construction of dual fluorescein reporter vector was achieved by the 3ʹ-UTR of wild type (WT) and mutant (MUT) GNA11 being cloned into pGL3-RB-REPORTTM. MiR-1249 mimic/inhibitor and PGL3-3ʹUTR-WT or PGL3-3ʹUTR-MUT were co-transfected into cells utilizing LipofectamineTM 2000 (Invitrogen). Finally, the luciferase activity was evaluated by using the Dual-Luciferase Reporter Assay System (Promega, Corporation).
Statistics
All results were executed with GraphPad Prism 7.0 and SPSS 22.0 software. Comparisons were carried out utilizing Student’s t-test (for single comparisons) or ANOVA test with Dunnett post hot (for multiple comparisons). Error bars indicated SD in figure legends. P < 0.01 was regarded as statistically significant.
Results
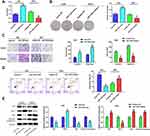
To inquire about the effects of miRNA on gastric cancer, we first searched the appropriate miRNA through bioinformatics analysis. The 13 common miRNAs (Table 2) were obtained from the intersection miRNAs that had meaningful survival (3273) and differential expression miRNAs (294) in gastric cancer that were acquired based on TCGA database (Figure 1A). To explore the function of miR-1249 on gastric cancer, we first analyzed miR-1249 expression level in gastric cancer tissues by using TCGA database. We discovered that the expression level of miR-1249 was highly expressed in gastric cancer tissues (Tumor, n=367) compared with that in paracancerous samples (Normal, n=13) (P=0.038, Figure 1B). Then, based on the cutoff value (3.01) of miR-1249 expression level, gastric cancer patients were divided into high (n=109) and low (n=109) expression groups. As presented in Figure 1C, high expression of miR-1249 was correlated with a worse prognosis in gastric cancer patients (P=0.016).
 |
Table 2 The 13 Predicted Common miRNAs |
To further probe the impacts of miR-1249 on gastric cancer, we assessed the properties of miR-1249 in gastric cancer cell lines AGS, MKN45 and SNU1. Normal human gastric cell line GES-1 was applied as a control. Figure 1D illustrates that miR-1249 expression was higher in all detected gastric cancer cell lines than that in GES-1 cell line. Moreover, SNU1 cell line showed a higher and AGS cell line exhibited a lower level of miR-1249 compared with other gastric cancer cell lines. Hence, AGS cell line was employed to up-regulate miR-1249 expression, whilst SNU1 cell line was used to down-regulate miR-1249 expression in the subsequent assays. Thus, AGS cells were transfected with miR-1249 mimic/mimic NC and SNU1 cells were transfected with miR-1249 inhibitor/inhibitor NC. Compared with miR-1249 mimic NC group, miR-1249 expression was accelerated by miR-1249 mimic in AGS cells, whereas miR-1249 inhibitor receded the miR-1249 level in SNU1 cells (Figure 1E). These consequences manifested that miR-1249 was highly expressed in gastric cancer tissues and cell lines, as well as might be concerned with poor prognosis for patients with gastric cancer.
Aberrant Expression of miR-1249 Influences the Proliferation, Motility and Apoptosis of Gastric Cancer Cells
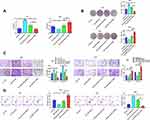
To determine the impacts of miR-1249 on gastric cancer cell proliferation, motility and apoptosis, CCK8, colony-formation, transwell and flow cytometry assays were employed. As shown in Figure 2A, the OD450 value of AGS cells fortified in miR-1249 mimic group relative to mimic NC group, proving that miR-1249 over-expressing facilitated AGS cell proliferation. On the contrary, the proliferation of SNU1 cells was reduced by miR-1249 down-regulation (Figure 2A). Moreover, the colony number of AGS cells was significantly increased by miR-1249 mimic compared with mimic NC group, while, the colony number of SNU1 cells was repressed by miR-1249 inhibitor when relative to inhibitor NC group (Figure 2B). Then, the transwell assay was used to seek the impacts of miR-1249 on the motility (migration and invasion) of gastric cancer cells. As presented in Figure 2C, miR-1249 up-regulation elevated the number of crystal violet-stained AGS cells. Inversely, miR-1249 ablation decreased the number of crystal violet-stained SNU1 cells (Figure 2C). Furthermore, to further explore the role of miR-1249 in gastric cancer, flow cytometric detection was utilized. Compared with the corresponding NC group, miR-1249 mimic restrained the apoptosis of AGS cells, whilst miR-1249 inhibitor promoted the apoptosis of SNU1 cells (Figure 2D). Besides, we also determined the protein levels of apoptosis-related proteins. Compared to the mimic NC group, miR-1249 mimic inhibited the protein levels of p53, Bax and Cleaved Caspase-9, while miR-1249 mimic facilitated the level of Bcl-2 in AGS cells (Figure 2E). However, miR-1249 inhibitor exhibited the opposite influences in SNU1 cells (Figure 2E). All the above findings demonstrated that miR-1249 over-expression presented an accelerative influence and miR-1249 depletion had a suppressive effect on gastric cancer cell proliferation and motility, while displayed an opposite impact on the apoptosis of gastric cancer cells.
GNA11 is a Target of miR-1249 and is Lowly Expressed in Human Gastric Cancer Tissues and Cell Lines
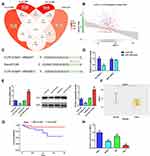
To probe the mechanisms of how miR-1249 affects the proliferation and motility of gastric cancer cells, we first predicted its potential target genes by bioinformatics analysis. There were three common genes (AIRE, VAMP2, GNA11) were obtained from the intersection of 3273 survival differences genes (ven_sur_allRNA), the 4428 expression difference genes (ven_dif_mRNA), the 4147 most related genes to hsa-miR-1249 through using the pearson correlation analysis (ven_cor_best) and the 3136 predicted target genes from the TargetScan software (target_gene), which all obtained the data from TCGA database (Figure 3A). Based on the analysis results and comprehensive literature review, GNA11 was chosen as one of the target genes of miR-1249 for further research. Then, we executed Pearson’s correlation analysis to ulteriorly analyze the relationship between miR-1249 and GNA11. As illustrated in Figure 3B, miR-1249 expression was negatively concerned with GNA11 expression (r=−0.170, P=0.006). Next, in order to further corroborate that GNA11 was a target gene of miR-1249, the luciferase assay was carried out. The seed region of GNA11 targeted by miR-1249 is displayed in Figure 3C. Besides, Figure 3D elucidates that the introduction of miR-1249 mimic weakened the luciferase activity of cells containing GNA11 3ʹ-UTR-WT instead of GNA11 3ʹ-UTR-MUT, compared with mimic NC group. qPCR and Western blotting analyses illustrated that miR-1249 over-expression resulted in a decrease in both mRNA and protein levels of GNA11, whereas miR-1249 depletion led to an increase in mRNA and protein levels of GNA11 (Figure 3E). Finally, we analyzed the expression of GNA11 in gastric cancer tissues and cell lines. As illustrated in Figure 3F, the expression level of GNA11 was lessened in gastric cancer tissues (n=357), compared to normal samples (n=23) (P=0.00011). According to the cutoff value (13.16) of GNA11 expression level, gastric cancer patients were fallen into high (n=106) and low (n=105) expression groups. Besides, low expression of GNA11 was concerned with worse survival (P=0.026), manifesting that GNA11 played a repressive effect in gastric cancer progression (Figure 3G). After that, we evaluated the impacts of GNA11 in gastric cancer cell lines AGS, MGC-803 and SNU1, GES-1 as a normal control. As shown in Figure 3H, GNA11 expression was lower in all detected gastric cancer cell lines than that in GES-1 cell line. The results in this chapter implied that miR-1249 directly targeted GNA11 to affect its expression on gastric cancer.
GNA11 is Involved in the Regulation of miR-1249 on Cell Growth, Migration and Invasion in Gastric Cancer
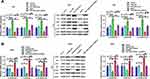
Moreover, rescue assays were performed to further probe whether GNA11 was associated with the regulation of miR-1249 on gastric cancer cell proliferation, motility and apoptosis. Through CCK8 and colony formation assays, we discovered that the promotive influence of miR-1249 over-expression on AGS cell proliferation was partly reversed by GNA11 over-expression (Figure 4A and B). Nevertheless, the introduction of GNA11 deficiency partially overcomes the repressive effect of the miR-1249 inhibitor on SNU1 cell proliferation (Figure 4A and B). Moreover, by transwell assays, we observed that the facilitative impact of miR-1249 mimic on AGS cell motility was counteracted by over-expression of GNA11 (Figure 4C). On the contrary, GNA11 silencing rescued the suppressive effect of miR-1249 down-regulation on SNU1 cell motility (Figure 4C). Additionally, through flow cytometry, we found that the restrained impact of miR-1249 high-expression on AGS cell apoptosis was inversed by GNA11 up-regulation (Figure 4D). However, the fortified effect of the miR-1249 inhibitor on the apoptosis of SNU1 cells was partially overcome by the stimulation of GNA11 depletion (Figure 4D). In summary, all the above data showed that miR-1249 could affect gastric cancer cell proliferation, motility and apoptosis through targeting GNA11.
The Influences of miR-1249/GNA11 on Gastric Cancer Cells is Partially Achieved by PI3K/AKT/mTOR Pathway
For the purpose of further examining the mechanisms of miR-1249/GNA11 on gastric cancer cell, we analyzed the expression changes of key factors in PI3K/AKT/mTOR signalling pathway through Western blotting. As displayed in Figure 5A, GNA11 over-expression reversed the acceerative impacts of miR-1249 up-regulation on the protein levels of p-PI3K/PI3K, p-AKT/AKT and p-mTOR/mTOR in AGS cells. However, the inhibitory influences of miR-1249 ablation on p-PI3K/PI3K, p-AKT/AKT and p-mTOR/mTOR protein levels expression levels were inversed by GNA11 knockdown in SNU1 cells (Figure 5B). These outcomes hinted that the properties of miR-1249/GNA11 on gastric cancer cells might be partially through PI3K/AKT/mTOR signalling pathway.
Discussion
In our study, we identified miR-1249 as an oncogene in gastric cancer for the first time. Besides, GNA11 was verified as one of the target genes of miR-1249 and was negatively regulated by miR-1249. Most importantly, the participation of miR-1249/GNA11 was proofed to be crucial in gastric cancer proliferation, motility and apoptosis, which was possibly achieved through regulating PI3K/AKT/mTOR pathway. Our findings highlighted miR-1249 as an oncogenic miRNA and GNA11 as a tumor suppressor in gastric cancer. Hence, miR-1249 and GNA11 might be defined as novel candidates for gastric cancer therapy.
MiRNAs were proved to participate in the progression of almost all vertebrate tissues, such as homeostasis, proliferation, cell cycle, motility, apoptosis and various pathological states.28 More importantly, abnormal miRNAs expression levels have facilitated or inhibited tumorigenesis through their ability to manage the expression of various non-coding and protein-coding genes.29 Recently, miR-1249 was recognized as a tumor-associated miRNA.30,31 As the study revealed by Fang et al,32 miR-1249 promoted the proliferation of glioblastoma cells by downregulating adenomatous polyposis coli 2 (APC2) expression. It has demonstrated that P53-induced miR-1249 repressed colorectal cancer cell growth, metastasis, and angiogenesis through targeting VEGFA and HMGA2.30 Kumar et al33 reported that circulating serum miR-1249-3p was differentially expressed in non-small cell lung cancer. Through regulating HNRNPK, miR-1249-3p knockdown inhibited the proliferation, colony formation, and invasion of hepatocellular carcinoma cells in vitro.20 In line with the above outcomes, our study displayed that miR-1249 was highly expressed in gastric cancer tissues and cell lines, and the participation of miR-1249 was testified to be crucial in gastric cancer proliferation, motility and apoptosis. Our findings offered a novel regulatory study on employing miR-1249 to modulate the cellular phenotypes of gastric cancer. Nevertheless, the mechanisms about the influences of miR-1249 in gastric cancer have rarely been studied and need further expound.
Herein, we uncovered GNA11 as a novel target gene of miR-1249 in gastric cancer. Furthermore, we proved that GNA11 was negatively regulated by miR-1249. It has been reported that activating Q209L/P mutations in GNA11 are present in various tumors.34 Studies have demonstrated that the impacts of GNA11 might be related to some cancers, such as thyroid cancer,35 melanoma,36 triple-negative breast cancer,34 and gastrointestinal stromal cancer27 etc. Murugan et al35 indicated the mutation status of GNA11 gene in thyroid cancer. By using bioinformatics analysis, GNA11 was identified as one of the molecular targets of triple-negative breast cancer34 or gastrointestinal stromal tumor.27 However, the impacts of GNA11 on gastric cancer have been really reported. Besides, the involved mechanisms of the effects of GNA11 on gastric cancer have not been well expounded. In our study, we discovered that GNA11 was lowly expressed in gastric cancer tissues and cell lines. Meanwhile, low expression of GNA11 was associated with a worse prognosis in gastric cancer patients. Especially, we also found that GNA11 suppressed the proliferation, motility but facilitated apoptosis of gastric cancer cells for the first time, implying that GNA11 is regarded as a tumor repressor in gastric cancer.
Moreover, the outcomes of the Western blotting analysis indicated that the influences of miR-1249/GNA11 on gastric cancer cell proliferation, motility and apoptosis might be partially through PI3K/AKT/mTOR pathway. Apoptosis is a key anti-tumour mechanism and several factors served as important effects in tumors by inducing apoptosis.37,38 Multiple signalling pathways are regulated to the activation of apoptosis, of which PI3K/AKT/mTOR is one of the most important signalling pathways.39,40 The PI3K/AKT/mTOR pathway regulates a number of malignant phenotypes, including anti-apoptotic, cell proliferation, growth, motility phenotypes, etc.41,42 miR-1249 was indicated to repress the growth and metastasis through regulating Akt/mTOR pathway in colorectal cancer.30 In a mutant GNA11-dependent manner, small molecules that repressing PI3K enhanced cell death of uveal melanoma.34 PI3K was regarded as one of the major pathways that influences the tumor suppressor gene GNA11 in colorectal cancer tissues.43 Consistent with the above studies, we discovered that the influences of miR-1249/GNA11 on gastric cancer cell properties might be partially through PI3K/AKT/mTOR pathway.
In summary, our findings manifested that miR-1249 was highly expressed and GNA11 was lowly expressed in gastric cancer tissues and cells. Meanwhile, the roles of miR-1249/GNA11 on gastric cancer cell proliferation, motility and apoptosis might be partially through PI3K/AKT/mTOR pathway, disclosing that miR-1249/GNA11 could serve as potential therapeutic targets for gastric cancer. The focus of our future work will use the in vivo experiments to further explore the biological effects of miR-1249 and GNA11 in gastric cancer.
Data Sharing Statement
The datasets used and analyzed during the current study are available from the corresponding author on reasonable request.
Disclosure
The authors declare that they have no conflicts of interest in this work.
References
1. Liang M, Huang G, Liu Z, et al. Elevated levels of hsa_circ_006100 in gastric cancer promote cell growth and metastasis via miR-195/GPRC5A signalling. Cell Prolif. 2019;52(5):e12661.
2. Strong VE. Progress in gastric cancer. Updates Surg. 2018;70(2):157–159.
3. Van Cutsem E, Sagaert X, Topal B, Haustermans K, Prenen H. Gastric cancer. Lancet. 2016;388(10060):2654–2664.
4. Bray F, Ferlay J, Soerjomataram I, Siegel RL, Torre LA, Jemal A. Global cancer statistics 2018: GLOBOCAN estimates of incidence and mortality worldwide for 36 cancers in 185 countries. CA Cancer J Clin. 2018;68(6):394–424.
5. Torre LA, Bray F, Siegel RL, Ferlay J, Lortet-Tieulent J, Jemal A. Global cancer statistics, 2012. CA Cancer J Clin. 2015;65(2):87–108.
6. Vineis P, Wild CP. Global cancer patterns: causes and prevention. Lancet. 2014;383(9916):549–557.
7. Song Z, Wu Y, Yang J, Yang D, Fang X. Progress in the treatment of advanced gastric cancer. Tumour Biol. 2017;39(7):1010428317714626.
8. Digklia A, Wagner AD. Advanced gastric cancer: current treatment landscape and future perspectives. World J Gastroenterol. 2016;22(8):2403–2414.
9. Tan Z. Recent advances in the surgical treatment of advanced gastric cancer: a review. Med Sci Monit. 2019;25:3537–3541.
10. Pang X, Zhou Z, Yu Z, et al. Foxo3a-dependent miR-633 regulates chemotherapeutic sensitivity in gastric cancer by targeting Fas-associated death domain. RNA Biol. 2019;16(2):233–248.
11. Link A, Kupcinskas J. MicroRNAs as non-invasive diagnostic biomarkers for gastric cancer: current insights and future perspectives. World J Gastroenterol. 2018;24(30):3313–3329.
12. Liu X, Cai H, Sheng W, Huang H, Long Z, Wang Y. microRNAs expression profile related with response to preoperative radiochemotherapy in patients with locally advanced gastric cancer. BMC Cancer. 2018;18(1):1048.
13. Zhang F, Li K, Yao X, et al. A miR-567-PIK3AP1-PI3K/AKT-c-Myc feedback loop regulates tumour growth and chemoresistance in gastric cancer. EBioMedicine. 2019;44:311–321.
14. Xiang S, Chen H, Luo X, et al. Isoliquiritigenin suppresses human melanoma growth by targeting miR-301b/LRIG1 signaling. J Exp Clin Cancer Res. 2018;37(1):184.
15. Liu B, Li J, Cairns MJ. Identifying miRNAs, targets and functions. Brief Bioinform. 2014;15(1):1–19.
16. Tutar Y. miRNA and cancer; computational and experimental approaches. Curr Pharm Biotechnol. 2014;15(5):429.
17. Armand-Labit V, Pradines A. Circulating cell-free microRNAs as clinical cancer biomarkers. Biomol Concepts. 2017;8(2):61–81.
18. Lai X, Eberhardt M, Schmitz U, Vera J. Systems biology-based investigation of cooperating microRNAs as monotherapy or adjuvant therapy in cancer. Nucleic Acids Res. 2019;47(15):7753–7766.
19. Cai M, Chen Q, Shen J, Lv C, Cai L. Epigenetic silenced miR-125a-5p could be self-activated through targeting Suv39H1 in gastric cancer. J Cell Mol Med. 2018;22(10):4721–4731.
20. Jia J, Zhan D, Li J, Li Z, Li H, Qian J. The contrary functions of lncRNA HOTAIR/miR-17-5p/PTEN axis and Shenqifuzheng injection on chemosensitivity of gastric cancer cells. J Cell Mol Med. 2019;23(1):656–669.
21. Dang W, Zhu Z. MicroRNA-1249 targets four-jointed box kinase 1 and reduces cell proliferation, migration and invasion of colon adenocarcinoma. J Gene Med. 2020;e3183.
22. Ye Y, Wei Y, Xu Y, et al. Induced MiR-1249 expression by aberrant activation of Hedegehog signaling pathway in hepatocellular carcinoma. Exp Cell Res. 2017;355(1):9–17.
23. Paraskevi A, Theodoropoulos G, Papaconstantinou I, Mantzaris G, Nikiteas N, Gazouli M. Circulating MicroRNA in inflammatory bowel disease. J Crohns Colitis. 2012;6(9):900–904.
24. Jiang ZH, Tang YZ, Song HN, Yang M, Li B, Ni CL. miRNA342 suppresses renal interstitial fibrosis in diabetic nephropathy by targeting SOX6. Int J Mol Med. 2019.
25. Schneider B, Riedel K, Zhivov A, et al. Frequent and yet unreported GNAQ and GNA11 mutations are found in uveal melanomas. Pathol Oncol Res. 2019;25(4):1319–1325.
26. Van Raamsdonk CD, Griewank KG, Crosby MB, et al. Mutations in GNA11 in uveal melanoma. N Engl J Med. 2010;363(23):2191–2199.
27. Park J, Yoo HM, Sul HJ, Shin S, Lee SW, Kim JG. Genetic characterization of molecular targets in Korean patients with gastrointestinal stromal tumors. J Gastric Cancer. 2020;20(1):29–40.
28. Sandiford OA, Moore CA, Du J, et al. Human aging and cancer: role of miRNA in tumor microenvironment. Adv Exp Med Biol. 2018;1056:137–152.
29. Hata A, Lieberman J. Dysregulation of microRNA biogenesis and gene silencing in cancer. Sci Signal. 2015;8(368):re3.
30. Chen X, Zeng K, Xu M, et al. P53-induced miR-1249 inhibits tumor growth, metastasis, and angiogenesis by targeting VEGFA and HMGA2. Cell Death Dis. 2019;10(2):131.
31. Shu H, Hu J, Deng H. miR-1249-3p accelerates the malignancy phenotype of hepatocellular carcinoma by directly targeting HNRNPK. Mol Genet Genomic Med. 2019;7(10):e00867.
32. Fang B, Li G, Xu C, Hui Y. MicroRNA miR-1249 downregulates adenomatous polyposis coli 2 expression and promotes glioma cells proliferation. Am J Transl Res. 2018;10(5):1324–1336.
33. Kumar S, Sharawat SK, Ali A, et al. Differential expression of circulating serum miR-1249-3p, miR-3195, and miR-3692-3p in non-small cell lung cancer. Hum Cell. 2020;33(3):839–849.
34. Lin FM, Yost SE, Wen W, et al. Differential gene expression and AKT targeting in triple negative breast cancer. Oncotarget. 2019;10(43):4356–4368.
35. Murugan AK, Yang C, Xing M. Mutational analysis of the GNA11, MMP27, FGD1, TRRAP and GRM3 genes in thyroid cancer. Oncol Lett. 2013;6(2):437–441.
36. Johnson DB, Roszik J, Shoushtari AN, et al. Comparative analysis of the GNAQ, GNA11, SF3B1, and EIF1AX driver mutations in melanoma and across the cancer spectrum. Pigment Cell Melanoma Res. 2016;29(4):470–473.
37. Ucker DS, Levine JS. Exploitation of Apoptotic Regulation in Cancer. Front Immunol. 2018;9:241.
38. Sethi G, Shanmugam MK, Warrier S, et al. Pro-apoptotic and anti-cancer properties of diosgenin: a comprehensive and critical review. Nutrients. 2018;10(5).
39. Kim MY, Kruger AJ, Jeong JY, et al. Combination therapy with a PI3K/mTOR dual inhibitor and chloroquine enhances synergistic apoptotic cell death in epstein-barr virus-infected gastric cancer cells. Mol Cells. 2019;42(6):448–459.
40. Kim J, Jung KH, Ryu HW, Kim DY, Oh SR, Hong SS. Apoptotic effects of Xanthium strumarium via PI3K/AKT/mTOR pathway in hepatocellular carcinoma. Evid Based Complement Alternat Med. 2019;2019:2176701.
41. De Santis MC, Porporato PE, Martini M, Morandi A. Signaling pathways regulating redox balance in cancer metabolism. Front Oncol. 2018;8:126.
42. Yu JS, Cui W. Proliferation, survival and metabolism: the role of PI3K/AKT/mTOR signalling in pluripotency and cell fate determination. Development. 2016;143(17):3050–3060.
43. Slattery ML, Herrick JS, Mullany LE, et al. The co-regulatory networks of tumor suppressor genes, oncogenes, and miRNAs in colorectal cancer. Genes Chromosomes Cancer. 2017;56(11):769–787.
 © 2021 The Author(s). This work is published and licensed by Dove Medical Press Limited. The full terms of this license are available at https://www.dovepress.com/terms.php and incorporate the Creative Commons Attribution - Non Commercial (unported, v3.0) License.
By accessing the work you hereby accept the Terms. Non-commercial uses of the work are permitted without any further permission from Dove Medical Press Limited, provided the work is properly attributed. For permission for commercial use of this work, please see paragraphs 4.2 and 5 of our Terms.
© 2021 The Author(s). This work is published and licensed by Dove Medical Press Limited. The full terms of this license are available at https://www.dovepress.com/terms.php and incorporate the Creative Commons Attribution - Non Commercial (unported, v3.0) License.
By accessing the work you hereby accept the Terms. Non-commercial uses of the work are permitted without any further permission from Dove Medical Press Limited, provided the work is properly attributed. For permission for commercial use of this work, please see paragraphs 4.2 and 5 of our Terms.