Back to Journals » OncoTargets and Therapy » Volume 11
MicroRNA-1225-5p behaves as a tumor suppressor in human glioblastoma via targeting of IRS1
Authors Li D, Chi G, Chen Z, Jin X
Received 22 June 2018
Accepted for publication 4 September 2018
Published 28 September 2018 Volume 2018:11 Pages 6339—6350
DOI https://doi.org/10.2147/OTT.S178001
Checked for plagiarism Yes
Review by Single anonymous peer review
Peer reviewer comments 3
Editor who approved publication: Dr Faris Farassati
Dongyuan Li, Guonan Chi, Zhuo Chen, Xingyi Jin
First Department of Neurosurgery, China-Japan Union Hospital of Jilin University, Changchun, Jilin 130033, People’s Republic of China
Background: MicroRNAs (miRNAs) play an important role in cancer initiation, progression, and metastasis by directly regulating their target genes.
Materials and methods: In this study, we observed that the miR-1225-5p expression level in glioblastoma tissues was significantly lower as compared with that in normal brain tissues, and its low expression was significantly associated with histopathological grade and poor patient prognosis.
Results: Through establishing a miR-1225-5p overexpression glioblastoma cell line, we found that ectopic overexpression of miR-1225-5p inhibited the proliferation, migration, and invasion of glioblastoma cells in vitro. Moreover, the growth of a glioblastoma xenograft tumor was attenuated by overexpression of miR-1225-5p. Further integrative studies suggested that the insulin receptor substrate 1 (IRS1) was a direct functional target of miR-1225-5p in glioblastoma, and the mRNA and protein levels of IRS1 in six human glioblastoma cell lines (A172, SW1783, U87, LN-229, SW1088, and T98G) were significantly higher as compared with normal human astrocytes.
Conclusion: These results suggest that miR-1225-5p may be a novel candidate for glioblastoma therapy.
Keywords: microRNA, microRNA-1225-5p, glioblastoma, insulin receptor substrate 1, prognosis
Introduction
As one of the most aggressive and common malignancies of the central nervous system in humans, glioblastoma is characterized by rapid infiltrative growth and cellular heterogeneity.1,2 Patients with high-grade glioblastoma have worse outcomes and shorter survival, with a median overall survival time of only 14.6 months as compared with patients who have low-grade tumors.3,4 Complete removal of glioblastoma tumor cells infiltrating or migrating to surrounding brain tissues by surgery is almost impossible. Besides surgery, radiotherapy and chemotherapy remain the mainstay of treatment for high-grade astrocytoma. However, due to the high invasiveness of glioblastoma cells and resistance to treatment, tumor recurrence and progression occur in >90% of glioblastoma patients shortly after surgery.5–8 Therefore, understanding the molecular basis and identifying the key molecule(s) responsible for the malignant phenotype of glioblastoma will help to identify potentially applicable therapeutic agents, and improve the therapeutic efficacy of the management of glioblastoma patients.
MicroRNAs (miRNAs) are a group of evolutionarily conserved, small, endogenous, non-coding RNAs that play important roles in the suppression of translation or induction of mRNA degradation as post-transcriptional gene regulators, by directly pairing and binding with the 3′-untranslated regions (UTRs) of specific target messenger RNAs (mRNAs).9–11 Accumulating evidence suggests that miRNAs are involved in a number of diverse physiological and pathological processes, particularly cancer pathogenesis, by directly targeting key oncogenes or tumor suppressors.12–16 It has been reported that miR-1225-5p is downregulated in human gastric carcinoma, and inhibits the proliferation and metastasis of gastric carcinoma cells.17 However, whether miR-1225-5p plays a role in glioblastoma remains largely unknown.
In the present study, we attempted to determine the role of miR-1225-5p in human glioblastoma. We observed that the downregulation of miR-1225-5p in glioblastoma was significantly associated with histopathological grade and poor patient prognosis. Mechanistic investigations revealed that miR-1225-5p inhibits the proliferation, migration, and invasion of glioblastoma cells in vitro. Moreover, the growth of glioblastoma xenograft tumors was attenuated by overexpression of miR-1225-5p. In addition, the insulin receptor substrate 1 (IRS1) was confirmed to be a direct functional target of miR-1225-5p in glioblastoma.
Materials and methods
Cell culture
Normal human astrocytes and six human glioblastoma cell lines (A172, SW1783, U87, LN-229, SW1088, and T98G) were purchased from the Type Culture Collection of the Chinese Academy of Sciences (Shanghai, People’s Republic of China). Apart from the astrocytes, which were cultured in Prigrow IV medium (ABM, Richmond, BC, Canada), all the other cells were maintained in DMEM (Gibco, Shanghai, People’s Republic of China) supplemented with 10% FBS, 100 U/mL penicillin, and 100 mg/mL streptomycin, and were incubated at 37°C in a humidified incubator supplemented with 5% CO2.
Clinical samples
A total of 44 glioma specimens and five normal brain samples frozen in liquid nitrogen were obtained from the China-Japan Union Hospital of Jilin University between January 2014 and May 2016. The study protocol was approved by the Ethics Review Committee of China-Japan Union Hospital of Jilin University, and written informed consent was obtained from all patients.
Generation of a cell line stably overexpressing miR-1225-5p (U87-1225)
The lentiviral plasmid pEZXMR03 (GeneCopoeia) expressing miR-1225-5p and Lenti-Pac HIV Expression Packaging mix (GeneCopoeia) were co-transfected into 293 T cells using Lipofectamine 2000 (Thermo Fisher Scientific, Waltham, MA, USA). After transfection for 48 hours, lentiviral particles were harvested and then transduced into the U87 glioblastoma cells, and the stably transfected cells were selected using puromycin and validated by reverse transcription PCR (RT-PCR).
Transfection and siRNA knockdown
SW1088 cells were transfected with siRNA sequence targeting IRS1 (AACCGGUUUCGAAAGAGA) using Lipofectamine 2000 (Invitrogen) according to the manufacturer’s instructions.
RNA extraction and quantitative RT-PCR
Total RNA was extracted from cultured cells or frozen tissues using the miRNeasy Mini kit (Qiagen, Hilden, Germany) and 1 mg RNA was reverse-transcribed using the miScript Reverse Transcription kit (Qiagen) for first-strand complementary DNA synthesis. Detection of miR-1225-5p and U6 utilized their specific primers separately. All the primers were synthesized by Invitrogen. The expression of miR-1225-5p was analyzed by quantitative PCR (qPCR) using the TaqMan microRNA assay kit (Applied Biosystems, Life Technologies).
Western blotting analysis
Western blotting was conducted according to the standard protocol with primary antibodies against IRS1, p-AKT, and AKT, and with β-catenin used as an internal control. All primary antibodies were purchased from Cell Signaling Technology, Inc. (Beverly, MA, USA).
Cell proliferation and colony-formation assay
Cell proliferation was detected using the MTT assay, according to the manufacturer’s instructions. Briefly, the cells were seeded in 96-well culture plates and incubated at 37°C with 5% CO2 for 3 days. At each designated time point, 10 μL MTT was added to the medium and then incubated at 37°C for 1 hour. Cell numbers were estimated by measuring the absorbance at 450 nm using a 96-well plate reader. For the colony-formation assay, 5,000 cells were plated in a six-well plate for 12 days. Colonies were subjected to methanol/acetone (1:1) fixation and stained with crystal violet.
Cell invasion assays
Cell invasion was assessed using Transwell chambers (24-well inserts; 8 μm-pore size; Millipore, Billerica, MA, USA) that had been pre-coated with Matrigel (200 μg/mL; BD Biosciences, Franklin Lakes, NJ, USA). Briefly, cells were added into the upper chamber and complete DMEM with 20% FBS was placed in the lower chamber. After 24 hours of incubation at 37°C with 5% CO2, the non-invading cells on the upper chamber were removed by wiping with a cotton swab. The cells that had infiltrated from Matrigel into the pores of the inserted filter were fixed and stained. The cell number in five randomly selected fields of view was visualized using a light microscope and counted.
Luciferase reporter assay
The 3′-UTR sequence of IRS1 was predicted to interact with miR-1225-5p, or a mutated sequence within the predicted target sites was synthesized and inserted into the XbaI and FseI sites of the pGL3 control luciferase reporter vector (Promega, Madison, WI, USA). Luciferase activity was measured 48 hours post-transfection using the Dual-Luciferase Reporter Assay system, according to the manufacturer’s instructions (Promega). For each sample, Renilla luciferase activity was normalized to firefly luciferase expression.
Xenograft assays in nude mice
All animal experiments were approved by the China-Japan Union Hospital of Jilin University Animal Care and Use Committee and followed up with the United Kingdom Coordinating Committee for Cancer Research (UKCCCR) with regard to the welfare of the animals. A total of 2×106 U87 or U87-1225 cells were injected subcutaneously into the dorsal flank of BALB/c nude mice. Six mice were included in one experimental group. The tumor diameters were measured every 3 days, and tumor volumes were calculated (width2×length×0.5). The mice were sacrificed 22 days after tumor implantation.
Statistical analysis
Statistical analyses were conducted using GraphPad Prism IV software. The Student’s t-test was used to analyze the results expressed as mean ± SD. The survival curves were plotted using Kaplan–Meier analysis. Differences were considered to be statistically significant at P<0.05.
Results
miR-1225-5p is downregulated in glioblastoma cell lines and tissues
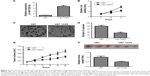
We first analyzed the expression of miR-1225-5p in six human glioblastoma cell lines (A172, SW1783, U87, LN-229, SW1088, and T98G). Compared with normal human astrocytes, the expression level of miR-1225-5p in these glioblastoma cell lines was significantly lower (Figure 1A). We further investigated the expression of miR-1225-5p in 44 glioblastoma samples (Table 1) and five normal brain tissue samples. The expression level of miR-1225-5p in the glioblastoma samples was also found to be significantly lower as compared with that in normal brain tissues (Figure 1B). Of note, the expression level of miR-1225-5p in advanced-stage (III and IV) glioblastoma was markedly lower when compared with that in earlier-stage (I and II) samples (Figure 1C).
  | Table 1 Clinicopathological features of 44 patients with glioblastoma |
Downregulation of miR-1225-5p is associated with disease progression and poor prognosis of patients with glioblastoma
To further investigate whether downregulation of miR-1225-5p is associated with glioblastoma progression and poor prognosis, the 44 abovementioned patients were divided into two groups – namely, low- and high-level expression of miR-1225-5p, based on the median value of the miR-1225-5p expression level among all patients (Table 2). Although the correlation between the miR-1225-5p expression level and the gender or age of the patients was not significant, the level of miR-1225-5p was negatively correlated with histopathological grade. Furthermore, as shown in Figure 1D as well as Figure S1A and B, the overall survival time of low miR-1225-5p expression patients was significantly shorter compared with that of the high miR-1225-5p expression group. Thus, miR-1225-5p suppresses the proliferation of glioblastoma cells in vitro and in vivo.
  | Table 2 Correlation between miR-1225-5p expression level and clinicopathological characteristics in 44 patients with glioblastoma |
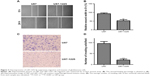
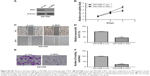
To investigate the role of miR-1225-5p in glioblastoma, we generated a glioblastoma cell line (U87-1225) stably overexpressing miR-1225-5p using retroviral vectors. As shown in Figure 2A, the miR-1225-5p expression level in U87-1225 cells was seven-fold higher when compared with that in parental U87 cells. The MTT assay demonstrated that overexpression of miR-1225-5p significantly suppressed the proliferation of U87 glioblastoma cells (Figure 2B). Furthermore, after being cultured for 14 days, the number of colonies in the U87-1225 group was significantly lower when compared with that in parental U87 cells (Figure 2C and D). Similar results were found in another glioblastoma cell line SW1088 (Figure S2A and D). The overexpression of miR-1225-5p significantly suppressed proliferation and colony formation of SW1088 cells. Moreover, miR-1225-5p significantly inhibited xenograft tumor growth in nude mice (Figure 2E and F).
miR-1225-5p suppresses the migration and invasion of glioblastoma cells
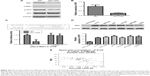
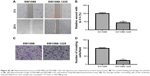
To further investigate the role of miR-1225-5p in glioblastoma, we next examined the effect of miR-1225-5p on the migration and invasion of U87 and SW1088 glioblastoma cell. In the scratch assay, overexpression of miR-1225-5p significantly attenuated wound healing (Figure 3A and B, Figure S3A and B). Moreover, we observed that overexpression of miR-1225-5p significantly abrogated the invasive ability of glioblastoma cells (Figure 3C and D, Figure S3C and D).
IRS1 is a direct target of miR-1225-5p in glioblastoma
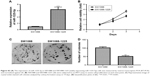
To investigate the mechanism underlying the inhibitory effect of miR-1225-5p on the proliferation, migration, and invasion of glioblastoma cells, the activation of the PI3K/AKT pathway in U87 cells was evaluated by Western blotting. We found that the overexpression of miR-1225-5p reduced both the expression and phosphorylation level of IRS1 and the phosphorylation of AKT, whereas it did not affect the total AKT expression (Figure 4A). Further, RT-PCR analysis demonstrated that overexpression of miR-1225-5p decreased the IRS1 mRNA level (Figure 4B). It was previously suggested that IRS1 is a direct target of miR-1225-5p in gastric carcinoma.17 As shown in Figure 4C, the relative luciferase activity of the pGL3-IRS1-3′-UTR-wt reporter was significantly inhibited by overexpression of miR-1225-5p; however, the luciferase activity of the pGL3-IRS1-3′-UTR-mutant was barely affected. Furthermore, the mRNA and protein levels of IRS1 in six human glioblastoma cell lines (A172, SW1783, U87, LN-229, SW1088, and T98G) was significantly higher when compared with that in normal human astrocytes (Figure 4D). Next, we conducted an expression correlation analysis. We identified 63 glioblastoma samples in The Cancer Genome Atlas. We determined the correlation of miR-1225-5p with IRS1. Our results showed that IRS1 negatively correlated miR-1225-5p (Figure 4E). To confirm miR-1225-5p targeting IRS1 in glioblastoma cell, we knocked-down IRS1 in SW1088 cells (Figure S4A). As shown in Figure S4B–F, when compared to si con cells, knockdown IRS1 suppressed proliferation, migration, and invasion in SW1088 cells.
Discussion
Based on the efforts of research groups worldwide, the abnormal expression and related role of miRNAs have been well established in certain types of cancer, acting as either oncogenes or tumor suppressors.18–25 It is crucial to identify cancer-specific miRNAs and further elucidate their role in tumor cell proliferation, invasion, and metastasis through specific targets, in order to identify novel therapeutic targets.26,27 miR-1225-5p was originally identified as a differentially expressed miRNA in adult renal progenitor cells, and was suggested to regulate the stemness and repair capacity of progenitor cells by targeting CD133, PAX2, and TLR2.28 It was recently reported that frequent downregulation of miR-1225-5p in gastric carcinoma was strongly correlated with an aggressive phenotype and poor prognosis in patients with gastric carcinoma.17 However, the expression pattern and clinical relevance of miR-1225-5p in human glioblastoma remains largely unknown.
In the present study, we demonstrated that miR-1225-5p was significantly downregulated in six human glioblastoma cell lines and 44 glioblastoma tissue samples, as compared with normal brain tissues. We further analyzed the correlation between miRNA expression and clinicopathological characteristics. A robust negative association between low expression levels of miR-1225-5p and advanced clinical stage was confirmed in patients with glioblastoma. Moreover, we demonstrated that low miR-1225-5p expression in glioblastoma was associated with poorer overall survival. To the best of our knowledge, this is the first study to provide evidence that miR-1225-5p may play a crucial role in the initiation and development of glioblastoma, and potentially acts as a prognostic biomarker in glioblastoma.
High proliferative, migratory, and invasive abilities are common characteristics of tumor cells.29 To characterize the function of miR-1225-5p in human glioblastoma, we induced stable overexpression of miR-1225-5p in U87 human glioblastoma cells, in which a relatively low expression level of miR-1225-5p was originally found. Our results from the MTT and colony-formation assays demonstrated that ectopic miR-1225-5p overexpression impaired short-term as well as long-term cell proliferation. Further, an in vivo xenograft assay confirmed the inhibitory role of miR-1225-5p in glioblastoma cells. Moreover, overexpression of miR-1225-5p was found to impair the migration and invasion of glioblastoma cells. These data suggest that miR-1225-5p plays a tumor-suppressing role in glioblastoma, similar to its function in gastric carcinoma.17
The major principle of miRNA function is direct binding to the 3′-UTR of the downstream target of miRNAs, and then inhibition of target gene expression at both the mRNA and protein levels.9,30 Consistently with previous reports, we found that overexpression of miR-1225-5p was significantly correlated with decreased expression of IRS1, as well as phosphorylation of AKT. As a docking protein, IRS1 plays a key role in transmitting signals from the insulin and insulin-like growth factor-1 (IGF-1) receptors to intracellular pathways, such as the PI3K/Akt pathway.31 Previous studies have shown that IRS1 may act as an oncogene; is involved in cancer cell growth, proliferation, migration, invasion, and differentiation;32 and is highly expressed in pancreatic cancer,33 breast cancer,34 colorectal cancer,35 and glioblastoma.36 Further, our results demonstrated the opposite pattern of expression when compared with miR-1225-5p, as IRS1 expression was commonly elevated in the six human glioblastoma cell lines. More importantly, the effect of miR-1225-5p overexpression on the luciferase activity of pGL3-IRS1-3′-UTR-wt, but not the mutant construct, confirmed that IRS1 is a direct target of miR-1225-5p in glioblastoma. Because it was previously demonstrated that miR-1225-5p inhibits the proliferation and metastasis of gastric carcinoma through IRS1, we consider IRS1 to be the major target of miR-1225-5p in cancer initiation and progression. Given the multiple targets of miR-1225-5p, future research should focus on investigating other potential regulators of glioblastoma progression.
In summary, the present study provided evidence that miR-1225-5p is downregulated in glioblastoma tissues and cell lines, and its low expression is correlated with advanced WHO grade and shorter overall survival of glioblastoma patients. The tumor suppressor effect of miR-1225-5p overexpression in vitro and in vivo makes it interesting to investigate for further development of miR-1225-5p-based therapeutic strategies.
Disclosure
The authors report no conflicts of interest in this work.
References
Siegel RL, Miller KD, Jemal A. Cancer statistics, 2016. CA Cancer J Clin. 2016;66(1):7–30. | ||
Northcott PA. Cancer: Keeping it real to kill glioblastoma. Nature. 2017;547(7663):291–292. | ||
Meyer MA. Malignant gliomas in adults. N Engl J Med. 2008;359(17):1850. | ||
Miller TE, Liau BB, Wallace LC, et al. Transcription elongation factors represent in vivo cancer dependencies in glioblastoma. Nature. 2017;547(7663):355–359. | ||
Wong ET, Hess KR, Gleason MJ, et al. Outcomes and prognostic factors in recurrent glioma patients enrolled onto phase II clinical trials. J Clin Oncol. 1999;17(8):2572–2578. | ||
Carson KA, Grossman SA, Fisher JD, Shaw EG. Prognostic factors for survival in adult patients with recurrent glioma enrolled onto the new approaches to brain tumor therapy CNS consortium phase I and II clinical trials. J Clin Oncol. 2007;25(18):2601–2606. | ||
Balmaceda C, Peereboom D, Pannullo S, et al. Multi-institutional phase II study of temozolomide administered twice daily in the treatment of recurrent high-grade gliomas. Cancer. 2008;112(5):1139–1146. | ||
da Fonseca CO, Schwartsmann G, Fischer J, et al. Preliminary results from a phase I/II study of perillyl alcohol intranasal administration in adults with recurrent malignant gliomas. Surg Neurol. 2008;70(3):259–266. | ||
Ambros V. The functions of animal microRNAs. Nature. 2004;431(7006):350–355. | ||
Bartel DP. MicroRNAs: genomics, biogenesis, mechanism, and function. Cell. 2004;116(2):281–297. | ||
Bentwich I, Avniel A, Karov Y, et al. Identification of hundreds of conserved and nonconserved human microRNAs. Nat Genet. 2005;37(7):766–770. | ||
Zhang L, Huang J, Yang N, et al. microRNAs exhibit high frequency genomic alterations in human cancer. Proc Natl Acad Sci U S A. 2006;103(24):9136–9141. | ||
Kayani MUR, Kayani MA, Malik FA, Faryal R. Role of miRNAs in Breast Cancer. Asian Pac J Cancer P. 2011;12:3175–3180. | ||
Mulrane L, Mcgee SF, Gallagher WM, O’Connor DP. miRNA dysregulation in breast cancer. Cancer Res. 2013;73(22):6554–6562. | ||
Serpico D, Molino L, di Cosimo S. microRNAs in breast cancer development and treatment. Cancer Treat Rev. 2014;40(5):595–604. | ||
Hata A, Lieberman J. Dysregulation of microRNA biogenesis and gene silencing in cancer. Sci Signal. 2015;8(368):re3. | ||
Zheng H, Zhang F, Lin X, et al. MicroRNA-1225-5p inhibits proliferation and metastasis of gastric carcinoma through repressing insulin receptor substrate-1 and activation of β-catenin signaling. Oncotarget. 2016;7(4):4647–4663. | ||
Chen Y, Liu W, Chao T, et al. MicroRNA-21 down-regulates the expression of tumor suppressor PDCD4 in human glioblastoma cell T98G. Cancer Lett. 2008;272(2):197–205. | ||
Cho WJ, Shin JM, Kim JS, et al. miR-372 regulates cell cycle and apoptosis of ags human gastric cancer cell line through direct regulation of LATS2. Mol Cells. 2009;28(6):521–527. | ||
Hu J, Guo H, Li H, et al. MiR-145 Regulates Epithelial to Mesenchymal Transition of Breast Cancer Cells by Targeting Oct 4. PLoS One. 2012;7(9):e45965. | ||
Yue X, Wang P, Xu J, et al. MicroRNA-205 functions as a tumor suppressor in human glioblastoma cells by targeting VEGF-A. Oncol Rep. 2012;27(4):1200–1206. | ||
Zhang QQ, Xu H, Huang MB, et al. MicroRNA-195 plays a tumor-suppressor role in human glioblastoma cells by targeting signaling pathways involved in cellular proliferation and invasion. Neuro Oncol. 2012;14(3):278–287. | ||
Chiang CH, Hou MF, Hung WC. Up-regulation of miR-182 by β-catenin in breast cancer increases tumorigenicity and invasiveness by targeting the matrix metalloproteinase inhibitor RECK. Biochim Biophys Acta. 2013;1830(4):3067–3076. | ||
Huang BS, Luo QZ, Han Y, Li XB, Cao LJ, Wu LX. microRNA-223 promotes the growth and invasion of glioblastoma cells by targeting tumor suppressor PAX6. Oncol Rep. 2013;30(5):2263–2269. | ||
Xia X, Li Y, Wang W, et al. MicroRNA-1908 functions as a glioblastoma oncogene by suppressing PTEN tumor suppressor pathway. Mol Cancer. 2015;14:154. | ||
Martello G, Rosato A, Ferrari F, et al. A MicroRNA targeting dicer for metastasis control. Cell. 2010;141(7):1195–1207. | ||
Gandellini P, Profumo V, Folini M, Zaffaroni N. MicroRNAs as new therapeutic targets and tools in cancer. Expert Opin Ther Targets. 2011;15(3):265–279. | ||
Sallustio F, Serino G, Costantino V, et al. Correction: miR-1915 and miR-1225-5p Regulate the Expression of CD133, PAX2 and TLR2 in Adult Renal Progenitor Cells. PLoS One. 2015;10(5):e0128258. | ||
Krogan NJ, Lippman S, Agard DA, Ashworth A, Ideker T. The cancer cell map initiative: defining the hallmark networks of cancer. Mol Cell. 2015;58(4):690–698. | ||
Zamore PD, Haley B. Ribo-gnome: the big world of small RNAs. Science. 2005;309(5740):1519–1524. | ||
Lima MH, Ueno M, Thirone AC, Rocha EM, Carvalho CR, Saad MJ. Regulation of IRS-1/SHP2 interaction and AKT phosphorylation in animal models of insulin resistance. Endocrine. 2002;18(1):1–12. | ||
Baserga R. The contradictions of the insulin-like growth factor 1 receptor. Oncogene. 2000;19(49):5574–5581. | ||
Bergmann U, Funatomi H, Kornmann M, Beger HG, Korc M. Increased expression of insulin receptor substrate-1 in human pancreatic cancer. Biochem Biophys Res Commun. 1996;220(3):886–890. | ||
Zhang J, Du YY, Lin YF, et al. The cell growth suppressor, mir-126, targets IRS-1. Biochem Biophys Res Commun. 2008;377(1):136–140. | ||
Zhou Y, Feng X, Liu YL, et al. Down-regulation of miR-126 is associated with colorectal cancer cells proliferation, migration and invasion by targeting IRS-1 via the AKT and ERK1/2 signaling pathways. PLoS One. 2013;8(11):e81203. | ||
Luan Y, Zuo L, Zhang S, Wang G, Peng T. MicroRNA-126 acts as a tumor suppressor in glioma cells by targeting insulin receptor substrate 1 (IRS-1). Int J Clin Exp Pathol. 2015;8(9):10345–10354. |
Supplementary materials
 © 2018 The Author(s). This work is published and licensed by Dove Medical Press Limited. The full terms of this license are available at https://www.dovepress.com/terms.php and incorporate the Creative Commons Attribution - Non Commercial (unported, v3.0) License.
By accessing the work you hereby accept the Terms. Non-commercial uses of the work are permitted without any further permission from Dove Medical Press Limited, provided the work is properly attributed. For permission for commercial use of this work, please see paragraphs 4.2 and 5 of our Terms.
© 2018 The Author(s). This work is published and licensed by Dove Medical Press Limited. The full terms of this license are available at https://www.dovepress.com/terms.php and incorporate the Creative Commons Attribution - Non Commercial (unported, v3.0) License.
By accessing the work you hereby accept the Terms. Non-commercial uses of the work are permitted without any further permission from Dove Medical Press Limited, provided the work is properly attributed. For permission for commercial use of this work, please see paragraphs 4.2 and 5 of our Terms.