Back to Journals » Cancer Management and Research » Volume 13
microR-4449 Promotes Colorectal Cancer Cell Proliferation via Regulation of SOCS3 and Activation of STAT3 Signaling
Authors Yan Z, Hong S, Song Y, Bi M
Received 4 June 2020
Accepted for publication 21 October 2020
Published 7 April 2021 Volume 2021:13 Pages 3029—3039
DOI https://doi.org/10.2147/CMAR.S266153
Checked for plagiarism Yes
Review by Single anonymous peer review
Peer reviewer comments 4
Editor who approved publication: Professor Yong Teng
Zhenkun Yan,1 Sen Hong,2 Yumei Song,3 Miaomiao Bi4
1Department of Endoscopy Center, The Third Hospital of Jilin University, Changchun, Jilin, 130022, People’s Republic of China; 2Department of Colorectal and Anal Surgery, The First Hospital of Jilin University, Changchun, Jilin, 130022, People’s Republic of China; 3Department of Thoracic Oncology, Tumor Hospital of Jilin Province, Changchun, Jilin, 130022, People’s Republic of China; 4Department of Ophthalmology, The Third Hospital of Jilin University, Changchun, Jilin, 130022, People’s Republic of China
Correspondence: Sen Hong
Department of Colorectal and Anal Surgery, The First Hospital of Jilin University, Changchun, Jilin, 130022, People’s Republic of China
Email [email protected]
Zhenkun Yan
Department of Endoscopy Center, The Third Hospital of Jilin University, Changchun, Jilin, 130022, People’s Republic of China
Email [email protected]
Introduction: Dysregulation of microRNAs (miRNAs), which represented a critical level of gene expression modulation, regulated the development of colorectal cancer. However, the functions of numerous miRNAs remain unclear in colorectal cancer.
Methods: The microarray data of GSE115513 were retrieved; subsequently, the differentially expressed miRNAs between 411 colon tumors and 381 normal colon mucosa were analyzed. Real-time PCR (RT-qPCR) and bioinformatic analysis were applied to examine the expression of miR-4449 in collected colorectal tumors and published microarray data. The activity of signal transducer and activator of transcription 3 (STAT3) signaling was detected by Western blotting and RT-qPCR. Dual-Luciferase assay and bioinformatic analysis were used to confirm the interaction between suppressor of cytokine signaling 3 (SOCS3) and miR-4449. Loss of function and rescue assays were performed to study the involvement of miR-4449 and SOCS3 in cell proliferation and apoptosis of colorectal cancer.
Results: Herein, we identified miR-4449 as a novel upregulated miRNA in colorectal cancer. Our data suggested that miR-4449 downregulation blocked the proliferation of colorectal cancer cells accompanied with the elevation of cell apoptosis. Decreased expression of miR-4449 led to inactivation of STAT3 pathway as indicated by dephosphorylation of STAT3 and downregulation of STAT3 target genes, including vascular endothelial growth factor (VEGF), c-Myc, baculovirus inhibitor of apoptosis containing 5 (BIRC5). Furthermore, SOCS3, a negative regulator of STAT3 pathway, was found to be a target gene of miR-4449. The data also showed that the inactivation of STAT3 pathway by miR-4449 inhibitor was realized by targeting SOCS3. Moreover, the biological function of miR-4449 downregulation was reversed by SOCS3 knockdown in colorectal cancer cells.
Conclusion: The current study revealed that miR-4449 promoted cell proliferation of colorectal cancer and was a promising potential therapeutic target for colorectal cancer.
Keywords: STAT3, microR-4449, colorectal cancer, SOCS3
Introduction
Colorectal cancer accounts for approximately 10% of all cancer cases globally, with nearly 2 million patients being diagnosed with colorectal cancer in 2018.1 Investigations into the molecular mechanisms of colorectal cancer have provided several drug targets and biomarkers for early detection of the disease.2 However, due to the heterogenicity of colorectal cancer cells, the 5-year overall survival rate remains 60%.3,4 Patients diagnosed as early-stage (Stage I–II) colorectal cancer exhibit a 5-year overall survival rate at more than 70%.5 However, patients diagnosed at advanced stage (Stage IV) colorectal cancer exert the worst prognosis with 5-year overall survival rate at merely 15%.5
MicroRNAs (miRNAs) are those single-stranded transcripts with no protein-coding potential.6 MiRNAs are critical components of competing endogenous RNA network in cells.7 Via binding to complementary sites on mRNA 3ʹUTR, miRNAs promote mRNA degradation or inhibit mRNA translation to repress gene expression.8 Dysregulation of miRNAs results in abnormal expression of target genes and is associated with cancer development.9 Hundreds of cancer-associated miRNAs have been reported in colorectal cancer.10,11 For example, miR-193a-3p, which is downregulated in BRAF-mutated colorectal cancers, inhibits colorectal cancer cell proliferation.12 MiR-4449 is highly expressed in serum from patients with multiple myeloma compared with samples from healthy volunteers.13 However, the precise function of miR-4449 in colorectal cancer is not known.
Signal transducer and activator of transcription 3 (STAT3) is a member of STATs family of transcription factors.14 Accumulating evidences have suggested that STAT3 is critical for the development of inflammatory bowel diseases including colorectal cancer.15 Upon activation by inflammatory factors such as IL-6, STAT3 is phosphorylated and translocated from cell surface into nucleus to activate transcription of pro-proliferative genes, thus promoting colorectal cancer cell proliferation and resistance to cell apoptosis.16 The activity of STAT3 pathway is repressed by several tumor suppressors in cancer cells. For example, suppressor of cytokine signaling 3 (SOCS3) is one of the most well-known negative regulators of STAT3 signaling.17 The KIR domain of SOCS3 facilitates binding to gp130 receptor and blocks phosphorylation of STAT3.18 Chu et al report that SOCS3 is downregulated in colorectal tumors, moreover, forced overexpression of SOCS3 induces cell growth arrest and cell apoptosis via inactivation of STAT3 signaling in colorectal cancer cells.19 The progression of colorectal cancer is the constitutive activation of STAT3 signaling induced by downregulation of inhibitors.20
In the current study, the bioinformatic analysis indicated that miR-4449 was an upregulated miRNA, which was retrieved from previously published microarray data.21 We aimed to address the influence of miR-4449 on the development of colorectal cancer and the regulatory association between miR-4449 and STAT3 signaling in colorectal cancer cells.
Materials and Methods
Collection of Tissues
Human colorectal tumors and the adjacent normal tissues were collected from 50 patients (35 males and 15 females, aged 46–71 years) during surgery resection at The First Hospital of Jilin University between June 2017 and April 2019. Samples were immediately stored in liquid nitrogen. The protocol of this study is approved by the Institutional Review Board of The First Hospital of Jilin University (IRB no. JLU201706-12). All patients provided written informed consents.
Cell Lines
Normal colonic epithelial cell line NCM460 was obtained from the Cell Bank of Type Culture Collection of the Chinese Academy of Sciences (Shanghai, China). Colorectal cancer cell lines HCT116, SW480, and HT29 were purchased from the American Type Culture Collection (Manassas, VA). NCM460 and HT29 cells were cultured in RPMI1640 (Thermo Fisher Scientific, Waltham, MA). HCT116 and SW480 cells were cultured in DMEM (Thermo Fisher Scientific). The medium was supplemented with 10% FBS (HyClone; Thermo Fisher Scientific). Cells were placed in a 37 °C incubator with 5% CO2.
Western Blotting
p-STAT3 (Cat. 9145), STAT3 (Cat.12640) antibodies were products of CST (Beverly, CA). SOCS3 (Cat. 04–004) and β-actin (Cat. A1978) antibodies were bought from Sigma-Aldrich. HRP-labeled secondary antibody of rabbit (Cat. ab6721) and mouse (Cat. ab6789) were purchased from Abcam (Cambridge, UK). Protein lysates were acquired from cells by using RIPA lysis buffer (Beyotime, Shanghai, China). Proteins were separated by SDS-PAGE gels and transferred to PVDF membrane. The blots were developed with ECL Plus Western blotting detection reagents (Millipore; Merck, Darmstadt, Germany).
RT-qPCR
TRIzol reagent (Invitrogen) was used to extract total RNA from cells and tissue samples. First-stranded cDNA was obtained via reverse-transcription by M-MLV Reverse Transcriptase kit (Invitrogen). RT-qPCR reaction was performed using the SYBR Green Master Mix (Applied Biosystems, MA) on an Applied Biosystems 7900HT sequence detection system. Relative expression of gene was calculated by the 2−ΔΔCt method. β-Actin and U6-snRNA were internal controls for the mRNA and miRNA, respectively. VEGF, c-Myc and BIRC5 were target genes of STAT3 signaling.22 Primers used were as follows: stem-loop: 5ʹ-CTCAACTGGTGTCGTGGA
GTCGGCAATTCAGTTGAGTGCCTC-3ʹ; miR-4449-F: 5ʹ-TCGGCAGGCGTCCC
GGGGCTGC-3ʹ; miR-4449-R: 5ʹ-CTCAACTGGTGTCGTGGA-3ʹ; U6-F: 5ʹ-CTCG
CTTCGGCAGCACA-3ʹ; U6-R: 5ʹ-AACGCTTCACGAATTTGCGT-3ʹ; SOCS3-F: 5ʹ-CCTGCGCCTCAAGACCTTC-3ʹ; SOCS3-R: 5ʹ-GTCACTGCGCTCCAGTAGA
A-3ʹ; VEGF-F: 5ʹ-AGGGCAGAATCATCACGAAGT-3ʹ; VEGF-R: 5ʹ-AGGGTCTCGATTGGATGGCA-3ʹ; c-Myc-F: 5ʹ-GTCAAGAGGCGAACACAC
AAC-3ʹ; c-Myc-R: 5ʹ-TTGGACGGACAGGATGTATGC-3ʹ; BIRC5-F: 5ʹ-AGGACCACCGCATCTCTACAT-3ʹ: BIRC5-R: 5ʹ-AAGTCTGGCTCGTTCTCA
GTG-3ʹ; β-actin-F: 5ʹ-CATGTACGTTGCTATCCAGGC-3ʹ; β-actin-R: 5ʹ-CTCCTTA
ATGTCACGCACGAT-3ʹ.
Bioinformatic Analysis
The microarray data of GSE115513 were downloaded from NCBI-GEO database (https://www.ncbi.nlm.nih.gov/gds/) with the GEO2R tool. The top 250 differentially expressed miRNAs between 411 colon tumors and 381 normal colon mucosa were analyzed. Many miRNAs in the list have been verified as colorectal cancer–related miRNAs. We selected miR-4449 for the current study based on the fact that miR-4449 ranked high in the list, which has not been studied yet. The target genes of miR-4449 were predicted by TargetScan V7.2 software (http://www.targetscan.org/vert_72/). Candidate targets were uploaded into DAVID software for pathway enrichment analysis. SOCS3 was selected due to its involvement in STAT3 signaling.
Transfection
miR-NC, miR-4449 mimic and miR-4449 inhibitor were synthesized by GenePharma (Suzhou, China). The siRNA targeting SOCS3 (SOCS3 siRNA) and the negative control (control siRNA) were purchased from GeneScript (Nanjing, China). For transient transfection, miR-NC or miR-4449 mimic or miR-4449 inhibitor or siRNA was mixed with Lipofectamine RNAiMax (Invitrogen, Carlsbad, CA) in serum-free DMEM and then added into each well. The transfection efficiency was determined by Western blotting or RT-qPCR at the timepoint of 48 hours after transfection procedure.
Cell Proliferation and Apoptosis Assay
Cell proliferation was detected by a CCK-8 kit (Dojindo, Tokyo, Japan). Cells were seeded in each well of 96-well plates (2500 cells/well). The CCK-8 solution was added into culture medium every 24 hours. The mixture was maintained for 2 hours and then subjected to detection of absorbance at 450 nm.
For analysis of cell apoptosis, cells were harvested and suspended in Annexin-V binding buffer provided by Dead Cell Apoptosis Kit with Annexin V FITC and PI (Invitrogen). Cells were then stained with Annexin-V-FITC and PI. After that, cells were subjected to flow cytometry analysis. Cells labeled with FITC without PI were early-apoptotic cells. Cells labeled with FITC and PI were late-apoptotic cells.
Dual-Luciferase Reporter Assay
Cells were seeded in each well of 24-well plates (30,000 cells/well). The 3ʹUTR of SOCS3 was ligated into luciferase plasmid pmirGLO. SOCS3 3ʹUTR-M (mutant) was prepared by introducing mutations into pmirGLO-SOCS3 3ʹUTR. In brief, 50 ng plasmids in combination with 10 nM miR-NC or miR-4449 mimic was co-transfected into cells. 48 hours after transfection, Dual-Luciferase Reporter Assay System (Promega, Madison, WI) was used to detect the relative luciferase activity of each group.
Statistical Analysis
All statistical calculation was carried out by GraphPad Prism 6.0 software. All experiments were repeated thrice. The correlation between miR-4449 and SOCS3 expression was studied by the Pearson correlation analysis. Student’s t test was applied to compare the differences between the two groups. Differences among four groups were firstly analyzed by one-way ANOVA followed by Tukey’s test. P value less than 0.05 was statistically significant.
Results
miR-4449 Was Overexpressed in Colorectal Cancer
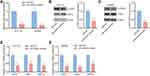
To screen colorectal cancer–related miRNAs, we retrieved microarray data containing colon tumors and normal colon mucosa from a previous study.21 In the list of significantly upregulated miRNAs, we found that miR-4449 was one of the highest upregulated miRNAs which has not been reported before. In 411 tumors, miR-4449 levels were nearly twofold higher than those of 381 normal tissues (Figure 1A). The miR-4449 expression was further evaluated in our collected 50 pairs of colorectal tumors and adjacent normal tissues. Consistently, miR-4449 was increased in colorectal tumors compared with their counterparts (Figure 1B). Moreover, we found that miR-4449 was highly expressed in tumors of advanced stage (Stage 4) compared with those of low stage (Stage 1) (Figure 1C). Thereafter, the expression of miR-4449 was detected in a normal colonic epithelial cell line NCM460 and a panel of colorectal cancer cell lines (HCT116, SW480 and HT29). Of interest, miR-4449 was significantly upregulated in colorectal cancer cell lines compared with NCM460 (Figure 1D).
Downregulation of miR-4449 Inactivated STAT3 Signaling
To further study the function of miR-4449 in colorectal cancer, we transfected miR-4449 inhibitor into colorectal cancer cells. In both HCT116 and SW480 cells, transfection of miR-4449 inhibitor decreased miR-4449 expression as indicated by RT-qPCR (Figure 2A). We next detected STAT3 and its active form (p-STAT3) in cells transfected with miR-4449 inhibitor. The Western blotting data showed that miR-4449 downregulation decreased p-STAT3 without affecting total STAT3 expression in HCT116 cells (Figure 2B), indicating that miR-4449 activated STAT3 signaling in colorectal cancer cells. Similar results were observed in SW480 cells (Figure 2C). As acknowledged, activated STAT3 enhanced transcription of cell proliferation–related genes such as VEGF, c-Myc and BIRC5. Via RT-qPCR, we found that miR-4449 downregulation decreased VEFG, c-Myc and BIRC5 mRNA expression in HCT116 and SW480 cells (Figure 2D–E), which further confirmed that miR-4449 regulated activation of STAT3 signaling in colorectal cancer.
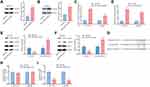
SOCS3 was a Potential Target of miR-4449
We used TargetScan software to predict targets of miR-4449. In the list of top 10 potential targets, it was found that SOCS3, a negative regulator of STAT3 signaling which prevents phosphorylation of STAT3 via binding to the catalytic domain of JAK,23 had putative binding site for miR-4449 (Figure 3A). In the samples, we found that SOCS3 was significantly downregulated in colorectal tumors compared with their counterparts (Figure 3B). Via Pearson correlation analysis, it was found that SOCS3 mRNA levels were negatively associated with miR-4449 levels in colorectal tumors (Figure 3C).
miR-4449 Directly Repressed SOCS3 Expression
We used Western blotting method to study the impact of miR-4449 on SOCS3 expression. As showed in Figure 4A and B, transfection of miR-4449 inhibitor increased SOCS3 protein expression in HCT116 and SW480 cells. Moreover, RT-qPCR showed that the mRNA levels of SOCS3 were also elevated upon miR-4449 downregulation in HCT116 and SW480 cells (Figure 4C). Next, we transfected miR-4449 mimic into HCT116 and SW480 cells to increase miR-4449 expression (Figure 4D). Western blotting showed that overexpression of miR-4449 decreased SOCS3 protein expression and elevated the ratio of p-STAT3/STAT3 in HCT116 and SW480 cells (Figure 4E and F). To further confirm the association between SOCS3 and miR-4449, we placed two point-mutations in putative binding site of miR-4449 in SOCS3 3ʹUTR (Figure 4G). In the Dual-Luciferase Reporter Assay, it was observed that miR-4449 mimic repressed the luciferase activity of SOCS3 3ʹUTR in HCT116 and SW480 cells (Figure 4H). Interestingly, miR-4449 mimic failed to alter the luciferase activity of SOCS3 3ʹUTR-M (Figure 4I), indicating that miR-4449 bound to 3ʹUTR of SOCS3 mRNA.
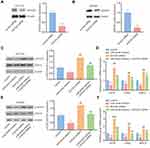
miR-4449 Regulated STAT3 Pathway via Regulating SOCS3
To examine the association among miR-4449, STAT3 pathway and SOCS3, we transfected SOCS3 siRNA into colorectal cancer cells. Transfection of SOCS3 siRNA decreased SOCS3 protein expression in HCT116 and SW480 cells (Figure 5A and B). As we expected, miR-4449 inhibitor decreased p-STAT3 expression, while SOCS3 siRNA increased p-STAT3 levels and reversed the effect of miR-4449 inhibitor on p-STAT3 in HCT116 cells (Figure 5C). Moreover, miR-4449 inhibitor repressed STAT3 target gene expression (VEGF, c-Myc, BIRC5), whereas SOCS3 siRNA increased the gene expression and attenuated the effect of miR-4449 inhibitor on STAT3 target gene expression (Figure 5D). Similar results were found in SW480 cells (Figure 5E and F).
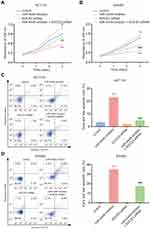
Downregulation of miR-4449 Negatively Regulated Cell Proliferation and Induced Cell Apoptosis in Colorectal Cancer Cells
STAT3 signaling is involved in colorectal cancer cell proliferation and resistance to cell apoptosis.24,25 Via the CCK-8 assay, we found that miR-4449 inhibitor repressed cell proliferation; however, SOCS3 siRNA promoted cell proliferation and reversed the effect of miR-4449 inhibitor on cell proliferation in HCT116 and SW480 cells (Figure 6A and B). With the flow cytometry, we observed that miR-4449 inhibitor evoked cell apoptosis, SOCS3 siRNA did not affect cell apoptosis but attenuated the effect of miR-4449 inhibitor on cell apoptosis in HCT116 and SW480 cells (Figure 6C and D).
Discussion
Slattery et al have identified many differentially expressed miRNAs between colorectal tumors and normal mucosa, such as miR-145-5p, miR-195-5p and miR-150-5p, by miRNA microarray. Cell-based experiments confirmed that many of these miRNAs were highly involved in the progression of colorectal cancer. For example, miR-145-5p regulated colorectal cancer cell proliferation, migration, invasion, stemness and resistance to radiotherapy via targeting SNAI1, PAK4 and TWIST1.26–28 In the present study, we re-analyzed the microarray data and found that miR-4449 was a novel dysregulated miRNA in colon cancer. Moreover, miR-4449 was dysregulated in multiple diseases, for instance, miR-4449 was one of the top 10 downregulated miRNAs in peripheral blood samples from patients with lung cancer.29 miR-4449 was highly expressed in serum from patients with multiple myeloma.12 However, there were few reports on the expression and function of miR-4449 in colorectal cancer. Currently, we detected miR-4449 expression in colorectal tumors and matched normal tissues, and found that miR-4449 was elevated in colorectal tumors especially in those of advanced stage, implying miR-4449 as a tumor-associated miRNA in colorectal cancer. In the future study, we will focus on the expression of miR-4449 in colorectal tumors from patients and normal colon tissues from patients without colorectal cancer to further examine the role of miR-4449.
JAK/STAT3 pathway is one of the most well-documented oncogenic signalings in colorectal cancer.30 Mechanistically, upon inflammatory stimulation, STAT3 is phosphorylated, translocated into nucleus and promotes gene expression.30,31 Constitutive activation of STAT3 pathway is a critical step for development of colorectal cancer.20,32 Accumulating evidences showed that various miRNAs could regulate STAT3 signaling in colorectal cancer at multiple levels. For example, downregulation of miR-198 increased expression of positive regulators of STAT3 pathway and promoted colorectal cancer cell proliferation.33,34 And miR-1299 directly repressed STAT3 expression in colorectal cancer.35 On the other hand, upregulation of several miRNAs activated STAT3 pathway via repressing SOCSs (negative regulators of STAT3 signaling) in colorectal cancer.36 Presently, we found that miR-4449 inhibitor inactivated STAT3 pathway in colorectal cancer cells.
SOCS3 could be regulated by multiple miRNAs. It has been reported that SOCS3 was targeted by miR-3473b, miR-203a-3p and miR-155 in cells of different origins.37–39 Our bioinformatic analysis revealed that SOCS3 3ʹUTR has complementary site for miR-4449. We also experimentally showed that miR-4449 directly targeted SOCS3 in colorectal cancer cells. More importantly, Western blotting and RT-qPCR showed that SOCS3 knockdown reversed the effect of miR-4449 inhibitor on STAT3 signaling. As acknowledged, sustained activation of STAT3 pathway promoted colorectal cancer cell proliferation and inhibited cell apoptosis.40,41 The data from CCK-8 and flow cytometry assays manifested that the miR-4449/SOCS3/STAT3 axis was pivotal for colorectal cancer cell proliferation and resistance to cell apoptosis.
In the future, we will establish stable miR-4449 knockdown colorectal cancer cells to further examine the role of miR-4449 in the initiation and development of colorectal cancer in several in vivo models.
Conclusions
Collectively, our finding suggested that miR-4449 was an upregulated miRNA in colorectal cancer and could activate STAT3 pathway via targeting SOCS3. miR-4449 may be a promising biomarker for patients with colorectal cancer.
Abbreviations
STAT3, signal transducers and activators of transcription 3; SOCS3, suppressor of cytokine signaling 3; CCK-8, Cell Counting Kit-8; VEGF, vascular endothelial growth factor; BIRC5, baculoviral IAP repeat containing 5.
Ethics Approval and Consent to Participate
All experiments were performed with patient consent. The protocol of this study is approved by the Institutional Review Board of The First Hospital of Jilin University (IRB no. JLU201706-12).
Disclosure
The authors declare that they have no competing interests in this work.
References
1. Bray F, Ferlay J, Soerjomataram I, Siegel RL, Torre LA, Jemal A. Global cancer statistics 2018: GLOBOCAN estimates of incidence and mortality worldwide for 36 cancers in 185 countries. CA Cancer J Clin. 2018;68(6):394–424. doi:10.3322/caac.21492
2. Yiu AJ, Yiu CY. Biomarkers in Colorectal Cancer. Anticancer Res. 2016;36(3):1093–1102.
3. Ji K, Zhang M, Chu Q, et al. The Role of p-STAT3 as a prognostic and clinicopathological marker in colorectal cancer: a systematic review and meta-analysis. PLoS One. 2016;11(8):e0160125. doi:10.1371/journal.pone.0160125
4. Dyson JK, Rutter MD. Colorectal cancer in inflammatory bowel disease: what is the real magnitude of the risk? World J Gastroenterol. 2012;18(29):3839–3848. doi:10.3748/wjg.v18.i29.3839
5. Cassia B. Impact of tumor location and variables associated with overall survival in patients with colorectal cancer: a Mayo Clinic Colon and Rectal Cancer Registry Study. Front Oncol. 2019;76(9). doi:10.3389/fonc.2019.00076
6. Mohr AM, Mott JL. Overview of microRNA biology. Semin Liver Dis. 2015;35(1):3–11. doi:10.1055/s-0034-1397344
7. Tay Y, Rinn J, Pandolfi PP. The multilayered complexity of ceRNA crosstalk and competition. Nature. 2014;505(7483):344–352. doi:10.1038/nature12986
8. Krol J, Loedige I, Filipowicz W. The widespread regulation of microRNA biogenesis, function and decay. Nat Rev Genet. 2010;11(9):597–610. doi:10.1038/nrg2843
9. Huang S, Tan X, Huang Z, Chen Z, Lin P, Fu SW. microRNA biomarkers in colorectal cancer liver metastasis. J Cancer. 2018;9(21):3867–3873. doi:10.7150/jca.28588
10. Tokarz P, Blasiak J. The role of microRNA in metastatic colorectal cancer and its significance in cancer prognosis and treatment.. Acta Biochim Pol. 2012;59(4):467–474. doi:10.18388/abp.2012_2079
11. Wei L, Wang X, Lv L, Zheng Y, Zhang N, Yang M. The emerging role of noncoding RNAs in colorectal cancer chemoresistance. Cellular Oncology. 2019;42(6):757–768. doi:10.1007/s13402-019-00466-8
12. Takahashi H, Takahashi M, Ohnuma S, et al. microRNA-193a-3p is specifically down-regulated and acts as a tumor suppressor in BRAF-mutated colorectal cancer. BMC Cancer. 2017;17(1):723. doi:10.1186/s12885-017-3739-x
13. Shen X, Ye Y, Qi J, et al. Identification of a novel microRNA, miR-4449, as a potential blood based marker in multiple myeloma. Clin Chem Lab Med. 2017;55(5):748–754. doi:10.1515/cclm-2015-1108
14. Furqan M, Akinleye A, Mukhi N, Mittal V, Chen Y, Liu D. STAT inhibitors for cancer therapy. J Hematol Oncol. 2013;6(1):90. doi:10.1186/1756-8722-6-90
15. Li Y, de Haar C, Chen M, et al. van der Woude CJ. Disease-related expression of the IL6/STAT3/SOCS3 signalling pathway in ulcerative colitis and ulcerative colitis-related carcinogenesis. Gut. 2010;59(2):227–235. doi:10.1136/gut.2009.184176
16. Zhong Z, Wen Z, Darnell JE. Stat3: a STAT family member activated by tyrosine phosphorylation in response to epidermal growth factor and interleukin-6. Science. 1994;264(5155):95–98. doi:10.1126/science.8140422
17. Inagaki-Ohara K, Kondo T, Ito M, Yoshimura A. SOCS, inflammation, and cancer. JAKSTAT. 2013;2(3):e24053. doi:10.4161/jkst.24053
18. Yoshimura A, Naka T, Kubo M. SOCS proteins, cytokine signalling and immune regulation. Nat Rev Immunol. 2007;7(6):454–465. doi:10.1038/nri2093
19. Chu Q, Shen D, He L, Wang H, Liu C, Zhang W. Prognostic significance of SOCS3 and its biological function in colorectal cancer. Gene. 2017;627:114–122. doi:10.1016/j.gene.2017.06.013
20. Grandis JR, Drenning SD, Zeng Q, et al. Constitutive activation of Stat3 signaling abrogates apoptosis in squamous cell carcinogenesis in vivo. Proc Natl Acad Sci U S A. 2000;97(8):4227–4232. doi:10.1073/pnas.97.8.4227
21. Slattery ML, Herrick JS, Pellatt DF, et al. MicroRNA profiles in colorectal carcinomas, adenomas and normal colonic mucosa: variations in miRNA expression and disease progression. Carcinogenesis. 2016;37(3):245–261. doi:10.1093/carcin/bgv249
22. Soutto M, Chen Z, Bhat AA, et al. Activation of STAT3 signaling is mediated by TFF1 silencing in gastric neoplasia. Nat Commun. 2019;10(1):3039. doi:10.1038/s41467-019-11011-4
23. Croker BA, Krebs DL, Zhang J-G, et al. SOCS3 negatively regulates IL-6 signaling in vivo. Nat Immunol. 2003;4(6):540–545. doi:10.1038/ni931
24. Zhang T. Overexpression of B7-H3 augments anti-apoptosis of colorectal cancer cells by Jak2-STAT3. World J Gastroenterol. 2015;21(6):1804–1813. doi:10.3748/wjg.v21.i6.1804
25. Zhang J, Lu Y, Yue X, et al. MiR-124 suppresses growth of human colorectal cancer by inhibiting STAT3. PLoS One. 2013;8(8):e70300. doi:10.1371/journal.pone.0070300
26. Zhu Y, Wang C, Becker SA, et al. miR-145 Antagonizes SNAI1-Mediated Stemness and Radiation Resistance in Colorectal Cancer. Mol Ther. 2018;26(3):744–754. doi:10.1016/j.ymthe.2017.12.023
27. Sheng N, Tan G, You W, et al. MiR-145 inhibits human colorectal cancer cell migration and invasion via PAK4-dependent pathway. Cancer Med. 2017;6(6):1331–1340. doi:10.1002/cam4.1029
28. Shen X, Jiang H, Chen Z, et al. MicroRNA-145 inhibits cell migration and invasion in colorectal cancer by targeting TWIST. Onco Targets Ther. 2019;12:10799–10809. doi:10.2147/OTT.S216147
29. He Q, Fang Y, Lu F, et al. Analysis of differential expression profile of miRNA in peripheral blood of patients with lung cancer. J Clin Lab Anal. 2019;33(9):e23003. doi:10.1002/jcla.23003
30. Zhang X, Hu F, Li G, et al. Human colorectal cancer-derived mesenchymal stem cells promote colorectal cancer progression through IL-6/JAK2/STAT3 signaling. Cell Death Dis. 2018;9(2):25. doi:10.1038/s41419-017-0176-3
31. Zhou J, Jiang Y, Zhao J, et al. Dp44mT, an iron chelator, suppresses growth and induces apoptosis via RORA-mediated NDRG2-IL6/JAK2/STAT3 signaling in glioma. Cellular Oncology. 2020;43(3):461–475. doi:10.1007/s13402-020-00502-y
32. Pan S, Deng Y, Fu J, et al. TRIM52 promotes colorectal cancer cell proliferation through the STAT3 signaling. Cancer Cell Int. 2019;19(1):57. doi:10.1186/s12935-019-0775-4
33. Li LX, Lam IH, Liang FF, et al. MiR-198 affects the proliferation and apoptosis of colorectal cancer through regulation of ADAM28/JAK-STAT signaling pathway. Eur Rev Med Pharmacol Sci. 2019;23(4):1487–1493. doi:10.26355/eurrev_201902_17106
34. Hong YG, Xin C, Zheng H, et al. miR-365a-3p regulates ADAM10-JAK-STAT signaling to suppress the growth and metastasis of colorectal cancer cells. J Cancer. 2020;11(12):3634–3644. doi:10.7150/jca.42731
35. Wang Y, Lu Z, Wang N, Zhang M, Zeng X, Zhao W. MicroRNA-1299 is a negative regulator of STAT3 in colon cancer. Oncol Rep. 2017;37(6):3227–3234. doi:10.3892/or.2017.5605
36. Ren D, Lin B, Zhang X, et al. Maintenance of cancer stemness by miR-196b-5p contributes to chemoresistance of colorectal cancer cells via activating STAT3 signaling pathway. Oncotarget. 2017;8(30):49807–49823. doi:10.18632/oncotarget.17971
37. Wang X, Chen S, Ni J, Cheng J, Jia J, Zhen X. miRNA-3473b contributes to neuroinflammation following cerebral ischemia. Cell Death Dis. 2018;9(1):11. doi:10.1038/s41419-017-0014-7
38. Xu JZ, Shao CC, Wang XJ, et al. circTADA2As suppress breast cancer progression and metastasis via targeting miR-203a-3p/SOCS3 axis. Cell Death Dis. 2019;10(3):175. doi:10.1038/s41419-019-1382-y
39. Li XD, Li XM, Gu JW, Sun XC. MiR-155 regulates lymphoma cell proliferation and apoptosis through targeting SOCS3/JAK-STAT3 signaling pathway. Eur Rev Med Pharmacol Sci. 2017;21(22):5153–5159. doi:10.26355/eurrev_201711_13832
40. Wang SW, Sun YM. The IL-6/JAK/STAT3 pathway: potential therapeutic strategies in treating colorectal cancer (Review). Int J Oncol. 2014;44(4):1032–1040. doi:10.3892/ijo.2014.2259
41. Cheng CC, Liao PN, Ho AS, et al. STAT3 exacerbates survival of cancer stem-like tumorspheres in EGFR-positive colorectal cancers: rNAseq analysis and therapeutic screening. J Biomed Sci. 2018;25(1):60. doi:10.1186/s12929-018-0456-y
 © 2021 The Author(s). This work is published and licensed by Dove Medical Press Limited. The full terms of this license are available at https://www.dovepress.com/terms.php and incorporate the Creative Commons Attribution - Non Commercial (unported, v3.0) License.
By accessing the work you hereby accept the Terms. Non-commercial uses of the work are permitted without any further permission from Dove Medical Press Limited, provided the work is properly attributed. For permission for commercial use of this work, please see paragraphs 4.2 and 5 of our Terms.
© 2021 The Author(s). This work is published and licensed by Dove Medical Press Limited. The full terms of this license are available at https://www.dovepress.com/terms.php and incorporate the Creative Commons Attribution - Non Commercial (unported, v3.0) License.
By accessing the work you hereby accept the Terms. Non-commercial uses of the work are permitted without any further permission from Dove Medical Press Limited, provided the work is properly attributed. For permission for commercial use of this work, please see paragraphs 4.2 and 5 of our Terms.