Back to Journals » OncoTargets and Therapy » Volume 7
Micro-ribonucleic acid expression profiling and bioinformatic target gene analyses in laryngeal carcinoma
Authors Lu Z, Lin Y, Jiang L, Chen L, Luo X, Song X, Chen S, Zhang S
Received 30 December 2013
Accepted for publication 1 February 2014
Published 5 April 2014 Volume 2014:7 Pages 525—533
DOI https://doi.org/10.2147/OTT.S59871
Checked for plagiarism Yes
Review by Single anonymous peer review
Peer reviewer comments 3
Zhong-Ming Lu,1 Ye-Feng Lin,1 Li Jiang,2 Liang-Si Chen,1 Xiao-Ning Luo,1 Xin-Han Song,1 Shao-Hua Chen,1 Si-Yi Zhang1
1Department of Otorhinolaryngology, 2Medical Research Center, Guangdong General Hospital and Guangdong Academy of Medical Sciences, Guangzhou, People's Republic of China
Abstract: Abnormal expression of micro-ribonucleic acid (miRNA) might be clinically valuable as a biomarker or treatment target in the early diagnosis, treatment, and prognosis of tumors. However, little is known concerning abnormal miRNA expression of laryngeal carcinoma, one of the most commonly encountered head and neck tumors. Microarray analysis was used to obtain miRNA-expression profiles of ten pairs of freshly frozen laryngeal carcinoma tissue and surrounding normal tissue specimens. Characteristic miRNAs that were significantly related to laryngeal carcinoma were identified. Verification was performed using an additional 32 pairs of samples. The expression of two miRNAs (miR-21-3p and miR-106b-3p) was upregulated in both microarray and quantitative real-time polymerase chain-reaction analyses, whereas the expression of six miRNAs (let-7f-5p, miR-10a-5p, miR-125a-5p, miR-144-3p, miR-195-5p, and miR-203) was downregulated. The decreased expression of let-7f-5p and miR-195-5p is a novel finding in head and neck cancer. The target genes of these miRNAs were also predicted through multiple software programs. The differential expression of miRNAs might be related to the early onset and development of laryngeal carcinoma, and may be exploited as new biomarkers and therapeutic targets in the treatment of laryngeal carcinoma.
Keywords: microRNA, laryngeal carcinoma, expression profiling, bioinformatics, target prediction
Introduction
Head and neck tumors constitute one of the main cancers endangering human health, ranking eighth in incidence among all cancer types. Laryngeal carcinoma is one of the most common types of head and neck tumors, and is the second-most prevalent of the respiratory tract tumors.1 Most laryngeal carcinoma cases are squamous cell carcinoma. The incidence of laryngeal carcinoma has been increasing in recent years. In the People’s Republic of China, the incidence of laryngeal carcinoma has increased by about 25% annually in an era of rapid industrialization.2,3 Although new surgical procedures, chemotherapeutic drugs, and more advanced radiotherapy have been applied to the treatment of laryngeal carcinoma in the past 30 years, the overall survival of patients with laryngeal carcinoma and other head and neck tumors alike has not improved, and in some cases has reduced.4 The survival rate of late-stage laryngeal carcinoma is only 30%–40%.5 Possible epidemiologically related factors for laryngeal carcinoma include smoking, drinking, air pollution, and occupational factors.6,7 The pathogenesis of laryngeal carcinoma is related to the activation of oncogenes, including BCL2 and c-MYC,8,9 and the inactivation of tumor-suppressor genes, including p53 and RB.10,11 However, the pathogenic mechanism of laryngeal carcinoma at the molecular level is still unclear, which has hindered therapeutic interventions and efforts to increase the survival rate.
Micro-ribonucleic acids (miRNAs) are small (19–25 nucleotides) noncoding RNAs.12 At the posttranscriptional level, miRNAs regulate the expression level of target genes, which influences the regulation of biological events, including individual development, apoptosis, proliferation, and differentiation.13,14 Recent studies have implicated miRNAs in the regulation of oncogenes or tumor-suppressor genes that are germane in the occurrence, development, invasion, and metastasis of a variety of tumors.15,16 Furthermore, miRNAs might be related to the pathogenesis of head and neck tumors, such as laryngeal carcinoma.17 The differential expression of miRNAs might have potential clinical value as biomarkers or treatment targets in the early diagnosis, treatment, and prognosis of laryngeal carcinoma.
Although miRNA-expression profiling of many head and neck tumors, including oral and thyroid carcinoma, has been reported,18,19 the situation is far less clear for laryngeal carcinoma, one of the most common head and neck tumors. The present study was undertaken to address this shortcoming. The aim of this study was to identify new diagnostic biomarkers for the early diagnosis of laryngeal carcinoma or potential treatment targets for laryngeal carcinoma.
Materials and methods
Tissue sampling and clinical data collection
A total of 42 pairs of surgically removed laryngeal carcinoma tissue and paracancerous tissue samples were collected from patients who received surgical resection treatment at Guangdong General Hospital from October 2007 to March 2009. Informed consent for tissue use was provided beforehand by all patients, and the study was approved by the ethics committee of Guangdong General Hospital. Laryngeal carcinoma was confirmed by pathological study. Normal laryngeal mucosa specimens were retrieved 10 mm outside the negative margin. Samples were placed in RNAlater TissueProtect Tubes (Qiagen, Hilden, Germany) and stored at -80°C. Postsurgical pathology confirmed the diagnosis of squamous cell carcinoma in all patients. The patients did not receive radio- or chemotherapy before surgery. Samples were randomly allocated into two groups for analysis using the miRNA microarray (ten pairs) or quantitative real-time polymerase chain reaction (qRT-PCR; 32 pairs).
RNA extraction
Total RNA was isolated using TRIzol® (Life Technologies, Carlsbad, CA, USA) and miRNeasy mini kit (Qiagen) according to the manufacturers’ instructions, which efficiently recovered all RNA species, including miRNAs. RNA quality and quantity was measured using an ND-1000 spectrophotometer (NanoDrop Technologies, Wilmington, DE, USA), and RNA integrity was determined by gel electrophoresis.
Microarray analysis of miRNA
Total RNA of ten pairs of laryngeal carcinoma tissue samples was randomly selected, and microarray analysis was completed with the sixth generation of miRCURY LNA™ (version 16.0) by Exiqon (Vedbaek, Denmark).
RNA labeling and array hybridization
After RNA isolation, the miRCURY Hy3™/Hy5™ power labeling kit (Exiqon) was used according to the manufacturer’s guidelines for miRNA labeling. After stopping the labeling procedure, the Hy3-labeled samples were hybridized on the miRCURY LNA array according to the instructions in the array manual. Following hybridization, the slides were washed several times using a wash buffer kit (Exiqon) and dried by centrifugation for 5 minutes at 400 rpm. The slides were scanned using a GenePix 4000B microarray scanner (Molecular Devices, Sunnyvale, CA, USA).
Data analyses
Scanned images were imported into the GenePix Pro 6.0 software (Molecular Devices) for grid alignment and data extraction. Hierarchical clustering was performed using MEV (multiexperiment viewer) version 4.6 (TIGR, Rockville, MD, USA).
Validation using qRT-PCR
Total RNA was extracted from 32 pairs of laryngeal carcinoma tissue and paracancerous tissue samples. The SYBR green assays (Life Technologies) with U6 as internal control was used, and qRT-PCR was performed to validate the results.
Reverse transcription
After total RNA extraction, complementary deoxyribonucleic acid was synthesized using 2 ng to 2 μg of total RNA by Moloney Murine Leukemia Virus Reverse Transcriptase (M-MLV RT) according to the manufacturer’s instructions (Life Technologies). Stem-loop reverse-transcriptase primer was added to total RNA (synthesized by RiboBio, Guangzhou, People’s Republic of China). The volume was expanded to 24 μL with ribonuclease (RNase)-free water and mixed well. After incubation at 70°C for 10 minutes, the mixture was placed in an ice bath for 2 minutes. The following reagents were added to the mixture in sequence: 2 μL of RNase inhibitor, 8 μL of 5× M-MLV RT buffer, 2 μL of M-MLV RT reverse transcriptase (RNase H-), and 2 μL of dNTP mixture (Life Technologies) and RNase-free water (TaKaRa Biotechnology, Dalian, People’s Republic of China) to a final volume of 40 μL. The mixture was placed at 30°C for 10 minutes, followed by 42°C incubation for 1 hour, and then the mixture was placed at 70°C for 15 minutes. The product of reverse transcription was used as the template of PCR.
qPCR
Amplification was performed on a fluorescent qPCR instrument (Bio-Rad Laboratories, Hercules, CA, USA). For each reaction, three duplicates were prepared. The total reaction volume was 20 μL, which included 10 μL of 2× Mix SYBR Green I (Life Technologies) fluorescent solution, 0.25 μL each of forward and reverse primers (10 μM), 1 μL of sample template, and RNase-free water that brought the volume to 20 μL. The reaction cycle was predenaturation at 95°C for 20 seconds, followed by 40 cycles of denaturation at 95°C for 10 seconds, annealing at 60°C for 20 seconds and extension at 70°C for 10 seconds. An amplification curve was plotted. A fluorescence signal was collected during the heating process from 60°C to 95°C after 40 cycles, and a melting curve was plotted. After each reaction, the threshold cycle (Ct) was calculated. Relative quantitation of gene expression was analyzed with the 2−ΔΔCt method using the obtained Ct values.
Statistical analyses
The results of the miRNA microarray were analyzed with significance analysis of microarrays (SAM) software. The q-value representing the false discovery rate (FDR) was set at 0.05, and miRNAs showing differential expression in laryngeal carcinoma tissue and paracancerous tissue were screened and identified. The results of qRT-PCR were processed with the 2−ΔΔCt method, and the difference between two groups was analyzed with the independent-samples t-test using SPSS 13.0 (SPSS, Chicago, IL, USA).
Results
Demographic and clinical data of the 42 laryngeal cancer patients
The results are summarized in Table 1.
  | Table 1 Demographic and clinical features of the laryngeal cancer patients |
Results of miRNA microarray
The miRNA microarray analysis screened out 780 miRNAs. After SAM analysis with FDR at 0.05, expressions of eleven miRNAs were upregulated in tumor tissue and 114 were downregulated, compared to expression in normal mucosa tissue (Tables 2, 3 and Figure 1).
  | Table 2 Significantly upregulated microRNAs (SAM, FDR ≤5%) |
  | Table 3 Significantly downregulated microRNAs (SAM, FDR ≤5%) |
qRT-PCR validation
Eight miRNAs (miR-21-3p, miR-106b-3p, let-7f-5p, miR-10a-5p, miR-125a-5p, miR-144-3p, miR-195-5p, and miR-203) were validated with qRT-PCR. The expression difference of these eight miRNAs was statistically significant between laryngeal carcinoma and normal mucosa tissue in the 32 pairs of specimens (P<0.05). Further comparison between the microarray and qRT-PCR results (Figure 2) showed that the expression pattern of these eight miRNAs in microarray was consistent with that in qRT-PCR.
Prediction of target genes
Potential target genes of the eight miRNAs were preliminarily predicted using bioinformatics software, including PicTar, miRanda, and TargetScan (Table 4 and Figure 3).
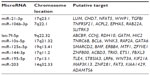  | Table 4 Putative target genes of differentially expressed micro-ribonucleic acids |
Discussion
In this study, the latest version of microarray mercury LNA (version 16.0) was used to analyze ten pairs of freshly frozen laryngeal carcinoma tissue and paracancerous tissue samples, and the miRNA-expression profile of laryngeal carcinoma was obtained. The results were verified by qRT-PCR using another 32 pairs of samples. The results between the two analyses were comparable. miR-21-3p and miR-106b-3p were upregulated, and let-7f-5p, miR-10a-5p, miR-125a-5p, miR-144-3p, miR-195-5p, and miR-203 were downregulated in the microarray and qRT-PCR analyses. The target genes of these miRNAs were also predicted through multiple software programs in this study.
Studies of miRNA-expression profiling of laryngeal carcinoma have suffered from some shortcomings. Most studies were conducted on head and neck squamous cell carcinoma,17,20,21 and few studies focused simply on laryngeal carcinoma.22 Although the disease locations of all kinds of tumors in the head and neck area are in close proximity, the larynx is part of the respiratory system, and so is different from such components of digestive system as the oral and pharyngeal areas.23,24 Laryngeal carcinoma has significantly distinctive pathological characteristics and prognosis from oral cancer, oropharyngeal cancer, and hypopharyngeal carcinoma. For example, the incidence of laryngeal carcinoma in men is significantly higher than that of other head and neck tumors.24 In addition, a comparative genomic hybridization study showed that the chromosomal patterns of laryngeal carcinoma and other head and neck squamous cell carcinomas were significantly different.25 Thus, without distinguishing laryngeal carcinoma from other head and neck squamous cell carcinoma, the results will be unavoidably biased.
In this study, the differentially expressed miRNAs detected in laryngeal carcinoma were different from reports of other head and neck tumors or laryngeal carcinoma. The numbers of differentially expressed miRNAs were larger, and were predominantly accounted for by downregulated miRNAs. The sample size of laryngeal carcinoma samples in this study was the largest among studies on head and neck tumors reported to date. Due to the heterogeneity of tumor samples, a large sample size might be beneficial for the accurate detection of abnormally expressed miRNA.26 In addition, the present study utilized the latest version of microarray chip with locked nucleic acid (LNA) technology,27 which guaranteed the maximal degree of reliability of the results. This is the first report on data from southern People’s Republic of China, and the different geographic locales of patients might also have contributed to the difference of the results. Furthermore, some studies only performed microarray detection without further verification by qRT-PCR, which has been recognized as a gold standard. This might also affect study results.
Among the eight differentially expressed miRNAs in laryngeal carcinoma verified by qRT-PCR, miR-21-3p,22 miR-106b-3p,20 miR-10a-5p,20 miR-125a-5p,17 miR-203,28 and miR-144-3p29 have been reported in other head and neck cancer studies. miR-125a-5p, miR-203, and miR-144-3p, which were only reported in other studies utilizing microarrays, were further verified by qRT-PCR in our study.
In addition, miRNAs not previously reported or inconsistent with previous head and neck squamous cell carcinoma or laryngeal carcinoma studies were also identified, and included let-7f-5p and miR-195-5p. Lethal-7 (let-7) was initially discovered in Caenorhabditis elegans. Being one of the first tumor-suppressive miRNAs identified, let-7 is downregulated in lung, ovarian, and breast cancer. Studies showed that let-7i is highly expressed17 in head and neck squamous cell carcinoma, and the expressions of let-7a, c, and e are low.20 In our study, let-7f-5p showed low expression, which was opposite to a report from Tran et al.30 The samples used in the prior study were from the cell lines of different head and neck squamous cell carcinomas, including tongue, tonsil, hypopharynx, and larynx, and no further verification of let-7f was conducted.30 Our study used freshly frozen laryngeal carcinoma tissue as samples, and qRT-PCR verification was performed. This further confirmed the advantage of singling out such diseases as laryngeal carcinoma, instead of studying a mixture of different types of head and neck tumors. miR-195 belongs to the miR-15/miR-16 family. miR-195 expression was downregulated in some tumor types, including hepatocellular carcinoma and breast cancer,31 suggesting that MIR195 might be an important tumor-suppressor gene. Also, it has been reported to be involved in the cell cycle, apoptosis, and proliferation through regulating such target genes as WEE1, CDK6, and BCL2.31 Our study is the first to demonstrate the possible presence of abnormally decreased expression of miR-195 in laryngeal carcinoma.
The molecular mechanism of miRNA in the regulation of laryngeal carcinoma-related genes is still unclear. Through such bioinformatic tools as PicTar, miRanda, and TargetScan,32,33 target genes of these miRNAs were preliminarily screened. The results showed that the target genes are those closely related to proliferation, cell-cycle regulation, and signal transduction, which are involved in aspects of tumorigenesis, tumor development, and metastasis. Bioinformatic computational methods for predicting target genes are necessary to guide experimental validation.
One limitation of this study was the lack of cell-function experiments and target-gene function validation. Whether the abnormally expressed miRNAs identified by microarray and verified by qRT-PCR are involved in the pathogenesis of laryngeal carcinoma requires further verification by function experiments.
In summary, miRNA-expression profiling of laryngeal carcinoma was performed using microarray analysis, and qRT-PCR verification was conducted. The decreased expression of let-7f-5p and miR-195-5p is a novel finding in head and neck cancer. In addition, target genes of identified miRNA were predicted using multiple databases. The identified miRNAs may have potential value as novel biomarkers and treatment targets of laryngeal carcinoma.
Acknowledgments
This work was supported by grants from the Guangdong Provincial Science and Technology Program 2011 (2011B031800148), the Second Batch Special Funds of Guangdong Provincial Finances Industrial Technology Research and Development 2011 (Z012011254), the Second Batch Science and Technology Program of Guangzhou Yuexiu District Science and Technology and Information Bureau 2012 (Z012012240), and the Special Care Project of the Guangdong Medical Science and Technology Research Fund 2013.
Disclosure
The authors report no conflicts of interest in this work.
References
Jemal A, Bray F, Center MM, Ferlay J, Ward E, Forman D. Global cancer statistics. CA Cancer J Clin. 2011;61(2):69–90. | |
Jaseviciene L, Gurevicius R, Obelenis V, Cicenas S, Juozulynas A. Trends in laryngeal cancer incidence in Lithuania: a future perspective. Int J Occup Med Environ Health. 2004;17(4):473–477. | |
Lu ST, Wei KR, Yu BH. Analysis of laryngeal cancer incidence rate in Zhongshan City in 1970–1999. Xian Dai Zhong Liu Yi Xue. 2004;12(2):158–160. | |
Hoffman HT, Porter K, Karnell LH, et al. Laryngeal cancer in the United States: changes in demographics, patterns of care, and survival. Laryngoscope. 2006;116(9 Pt 2 Suppl 111):1–13. | |
Gourin CG, Conger BT, Sheils WC, Bilodeau PA, Coleman TA, Porubsky ES. The effect of treatment on survival in patients with advanced laryngeal carcinoma. Laryngoscope. 2009;119(7):1312–1317. | |
Hashibe M, Boffetta P, Zaridze D, et al. Contribution of tobacco and alcohol to the high rates of squamous cell carcinoma of the supraglottis and glottis in Central Europe. Am J Epidemiol. 2007;165(7):814–820. | |
Shangina O, Brennan P, Szeszenia-Dabrowska N, et al. Occupational exposure and laryngeal and hypopharyngeal cancer risk in Central and Eastern Europe. Am J Epidemiol. 2006;164(4):367–375. | |
Marioni G, Staffieri A, Lionello M, et al. Relationship between anti-apoptotic proteins survivin and Bcl-2, and response to treatment in patients undergoing post-operative RT for laryngeal cancer: a pilot study. J Oral Pathol Med. 2013;42(4):339–344. | |
Fu S, Guo Y, Chen H, et al. MYCT1-TV, a novel MYCT1 transcript, is regulated by c-Myc and may participate in laryngeal carcinogenesis. PLoS One. 2011;6(10):e25648. | |
Jalali MM, Heidarzadeh A, Zavarei MJ, Sarmast H. p53 Overexpression impacts on the prognosis of laryngeal squamous cell carcinomas. Asian Pac J Cancer Prev. 2011;12(7):1731–1734. | |
Boonyaphiphat P, Pruegsanusak K, Thongsuksai P. The prognostic value of p53, Bcl-2 and Bax expression in laryngeal cancer. J Med Assoc Thai. 2012;95(10):1317–1320. | |
Fabbri M, Croce CM, Calin GA. MicroRNAs. Cancer J. 2008; 14(1):1–6. | |
Croce CM, Calin GA. miRNAs, cancer, and stem cell division. Cell. 2005;122(1):6–7. | |
Ambros V. The functions of animal microRNAs. Nature. 2004;431(7006):350–355. | |
Esquela-Kerscher A, Slack FJ. Oncomirs – microRNAs with a role in cancer. Nat Rev Cancer. 2006;6(4):259–269. | |
Calin GA, Croce CM. MicroRNA signatures in human cancers. Nat Rev Cancer. 2006;6(11):857–866. | |
Ramdas L, Giri U, Ashorn CL, et al. miRNA expression profiles in head and neck squamous cell carcinoma and adjacent normal tissue. Head Neck. 2009;31(5):642–654. | |
Gombos K, Horvath R, Szele E, et al. miRNA expression profiles of oral squamous cell carcinomas. Anticancer Res. 2013;33(4):1511–1517. | |
Wang Z, Zhang H, He L, et al. Association between the expression of four upregulated miRNAs and extrathyroidal invasion in papillary thyroid carcinoma. Onco Targets Ther. 2013;6:281–287. | |
Hui AB, Lenarduzzi M, Krushel T, et al. Comprehensive microRNA profiling for head and neck squamous cell carcinomas. Clin Cancer Res. 2010;16(4):1129–1139. | |
Chang SS, Jiang WW, Smith I, et al. MicroRNA alterations in head and neck squamous cell carcinoma. Int J Cancer. 2008;123(12):2791–2797. | |
Cao P, Zhou L, Zhang J, et al. Comprehensive expression profiling of microRNAs in laryngeal squamous cell carcinoma. Head Neck. 2013;35(5):720–728. | |
Almadori G, Bussu F, Paludettii G. Predictive factors of neck metastases in laryngeal squamous cell carcinoma. Towards an integrated clinico-molecular classification. Acta Otorhinolaryngol Ital. 2006;26(6):326–334. | |
Jemal A, Siegel R, Ward E, et al. Cancer statistics, 2006. CA Cancer J Clin. 2006;56(2):106–130. | |
Huang Q, Yu GP, McCormick SA, et al. Genetic differences detected by comparative genomic hybridization in head and neck squamous cell carcinomas from different tumor sites: construction of oncogenetic trees for tumor progression. Genes Chromosomes Cancer. 2002;34(2):224–233. | |
Chuaqui RF, Bonner RF, Best CJ, et al. Post-analysis follow-up and validation of microarray experiments. Nat Genet. 2002;32 Suppl:509–514. | |
Wang B, Howel P, Bruheim S, et al. Systematic evaluation of three microRNA profiling platforms: microarray, beads array, and quantitative real-time PCR array. PLoS One. 2011;6(2):e17167. | |
Zhong Q, Fang JG, Huang ZG, et al. Expression of miRNA in laryngeal squamous cell carcinoma tissues. Chin J Cancer Prev Treat. 2010;17(14):1073–1076. | |
Wang P, Fu T, Wang X, Zhu W. [Primary study of miRNA expression patterns in laryngeal carcinoma by microarray]. Lin Chung Er Bi Yan Hou Tou Jing Wai Ke Za Zhi. 2010;24(12):535–538. Chinese. | |
Tran N, McLean T, Zhang X, et al. MicroRNA expression profiles in head and neck cancer cell lines. Biochem Biophys Res Commun. 2007;358(1):12–17. | |
He JF, Luo YM, Wan XH, Jiang D. Biogenesis of MiRNA-195 and its role in biogenesis, the cell cycle, and apoptosis. J Biochem Mol Toxicol. 2011;25(6):404–408. | |
Witkos TM, Koscianska E, Krzyzosiak WJ. Practical aspects of microRNA target prediction. Curr Mol Med. 2011;11(2):93–109. | |
Tian Z, Greene AS, Pietrusz JL, Matus IR, Liang M. MicroRNA-target pairs in the rat kidney identified by microRNA microarray, proteomic, and bioinformatic analysis. Genome Res. 2008;18(3):404–411. |
 © 2014 The Author(s). This work is published and licensed by Dove Medical Press Limited. The full terms of this license are available at https://www.dovepress.com/terms.php and incorporate the Creative Commons Attribution - Non Commercial (unported, v3.0) License.
By accessing the work you hereby accept the Terms. Non-commercial uses of the work are permitted without any further permission from Dove Medical Press Limited, provided the work is properly attributed. For permission for commercial use of this work, please see paragraphs 4.2 and 5 of our Terms.
© 2014 The Author(s). This work is published and licensed by Dove Medical Press Limited. The full terms of this license are available at https://www.dovepress.com/terms.php and incorporate the Creative Commons Attribution - Non Commercial (unported, v3.0) License.
By accessing the work you hereby accept the Terms. Non-commercial uses of the work are permitted without any further permission from Dove Medical Press Limited, provided the work is properly attributed. For permission for commercial use of this work, please see paragraphs 4.2 and 5 of our Terms.



