Back to Journals » Neuropsychiatric Disease and Treatment » Volume 17
Methylation of the RIN3 Promoter is Associated with Transient Ischemic Stroke/Mild Ischemic Stroke with Early Cognitive Impairment
Authors Miao M , Yuan F, Ma X, Yang H, Gao X, Zhu Z , Bi J
Received 14 May 2021
Accepted for publication 23 July 2021
Published 10 August 2021 Volume 2021:17 Pages 2587—2598
DOI https://doi.org/10.2147/NDT.S320167
Checked for plagiarism Yes
Review by Single anonymous peer review
Peer reviewer comments 2
Editor who approved publication: Dr Taro Kishi
Meng Miao, 1 Fang Yuan, 2 Xiaotian Ma, 3 Haiming Yang, 1 Xiang Gao, 1 Zhengyu Zhu, 4 Jianzhong Bi 4
1Department of Neurology, Qilu Hospital (Qingdao), Cheeloo College of Medicine, Shandong University, Qingdao, Shandong, 266035, People’s Republic of China; 2Department of Health Care, Qilu Hospital (Qingdao), Cheeloo College of Medicine, Shandong University, Qingdao, Shandong, 266035, People’s Republic of China; 3Department of Medicine Experimental Center, Qilu Hospital (Qingdao), Cheeloo College of Medicine, Shandong University, Qingdao, Shandong, 266035, People’s Republic of China; 4Department of Neurology, The Second Hospital, Cheeloo College of Medicine, Shandong University, Jinan, Shandong, 250033, People’s Republic of China
Correspondence: Zhengyu Zhu; Jianzhong Bi
Department of Neurology, The Second Hospital, Cheeloo College of Medicine, Shandong University, Jinan, Shandong, 250033, People’s Republic of China
Tel +8617660083488
; +8615153178111
Email [email protected]; [email protected]
Background: Early cognitive impairment after transient ischemic stroke (TIA)/mild ischemic stroke (MIS) is common but easily overlooked. It has been demonstrated that DNA methylation plays a significant role in cognitive impairment and ischemic stroke. Furthermore, it has been reported that the RIN3 gene influences transportation of the amyloid β-protein. However, to our knowledge, there has been no research related to correlations between RIN3 methylation and early-onset cognitive impairment after TIA/MIS. Therefore, this study aimed to investigate this relationship in TIA/MIS patients.
Methods: This study include 28 control subjects and 84 patients with TIA/MIS who were evaluated within 7 days of TIA/MIS onset using four single-domain cognitive scales. In addition, DNA methylation of whole blood was tested. RIN3 methylation was compared between TIA/MIS and control groups and between TIA/MIS patients with early cognitive impairment and those without early cognitive impairment. Clinical variables and RIN3 methylation sites with statistical differences were then used to construct a predictive model.
Results: Hypomethylation of the RIN3 gene was observed in the whole blood of TIA/MIS patients relative to healthy controls. Furthermore, patients with early cognitive impairment after TIA/MIS had hypomethylation of RIN3 relative to those without early cognitive impairment.
Conclusion: RIN3 methylation is strongly associated with TIA/MIS and TIA/MIS with early cognitive impairment. It is possible to influence the disease process by methylation via appropriate lifestyle and clinical interventions, and methylation of RIN3 gene sites may predict the occurrence of TIA/MIS with early cognitive impairment.
Keywords: amyloid &Bgr;-protein, cognitive impairment, hypomethylation, ischemic stroke, transportation
Corrigendum for this paper has been published
Introduction
Ischemic stroke, one of the most common cerebral vascular diseases, can lead to neurological and cognitive impairment and can also accelerate cognitive disorders. Cognitive impairment can also occur in the early phase after a transient ischemic stroke (TIA) or a mild ischemic stroke (MIS). Since the dyskinetic symptoms of TIA/MIS are mild and short-lived, cognitive impairment due to TIA/MIS is easily overlooked, and the cognitive level of patients after onset of TIA/MIS is not routinely assessed in clinical practice. Currently, the specific pathogenesis of early cognitive impairment after TIA/MIS is unclear and requires further research.
It has been demonstrated that the incidence of early cognitive impairment after TIA/MIS can be as high as 32%–70%.1–4 Cognitive impairment is common in the acute phase of TIA/MIS and is more common within 7 days after TIA/MIS. Of note, the recovery of cognition in these patients does not parallel the resolution of somatic motor symptoms. Also, the acute phase of cognitive impairment is highly correlated with persistent future cognitive decline.2
DNA methylation, an important form of epigenetics, plays a significant role in several neurodegenerative pathologies, including Alzheimer’s disease (AD) and parkinsonism. Abnormal methylation of several genes has also been observed in ischemic cerebrovascular disease. The RIN3 gene, located on chromosome 14, acts as a stimulating factor to stabilise the transportation of GTP-RAB5 to endosomes at the plasma membrane. This process is associated with cellular endocytosis and has a negative effect on endocytosis of the amyloid β-protein (Aβ),5,6 which plays a role in the onset of both AD and vascular cognitive impairment (VCI). Studies have shown that the RIN3 gene is highly expressed in AD7 and that this gene is hypomethylated in the whole blood of patients with early-onset AD.8
Based on these previous studies, we hypothesised that abnormal expression of the RIN3 gene would impact the occurrence of cognitive impairment. Furthermore, we hypothesized that methylation, the most important manifestation of epigenetics, would affect expression of the RIN3 gene. To our knowledge, there is no existing research on the correlation between RIN3 gene methylation and early-onset cognitive impairment after TIA/MIS. Therefore, this study aimed to determine if hypomethylation of the RIN3 gene was associated with TIA/MIS with early cognitive impairment.
In this study, we examined whole-blood methylation levels in three groups, including TIA/MIS patients with early cognitive impairment, TIA/MIS patients without cognitive impairment, and non-infarcted control subjects, in order to investigate the correlation between whole-blood RIN3 methylation levels and TIA/MIS with early cognitive impairment. In addition, we aimed to identify significant predictive factors for construction of predictive models and for development of interventions.
Materials and Methods
Participants
A total of 84 patients (age < 75 years) with TIA/MIS were prospectively recruited from the Department of Neurology at the Qilu Hospital of Shandong University in Qingdao, China between June 2019 and July 2020. Mild stroke was defined as an acute cerebral infarction with a National Institutes of Health Stroke Scale (NIHSS) score of less than 5, and TIA was defined as a sudden focal neurological deficit in the brain, spinal cord, or retina lasting less than 24 h, which results from atherosclerotic vascular causes and is not associated with an acute cerebral infarction. All TIA/MISs were recorded using the Trial of Org 10172 in acute stroke treatment (TOAST) classification system and confirmed using cranial computed tomography (CT) and/or magnetic resonance imaging (MRI). Patients were evaluated within 7 days of TIA/MIS onset using four single-domain cognitive scales:9–16 the Boston Naming Test (BNT) for language (abnormality: ≤21.5 points; adjusted value: ≤19.5 points if the education period was ≤9 years and ≤21.5 points if the education period was >9 years), the Auditory Verbal Learning Test (AVLT) for memory (abnormality: <5 points after 5 min), and the Trail Making Test (TMT)-A for visuospatial function (abnormality: ≥98.5 s; adjusted value: ≥80.5 s for patients aged ≤64 years, ≥90.5 s for those aged 64−74 years, and ≥90.5 s for those aged ≥75 years, with a maximum time of 150 s), and TMT-B for executive function (abnormality: ≥188.5 s; adjusted value: ≥150.5 s for patients aged ≤64 years, ≥165.5 s for those aged 64−74 years, and ≥199.5 s for those aged ≥75 years, with a maximum time of 300 s). Cognitive impairment was determined by more than one (≥1) scale that has abnormal results. All cognitive screening were performed by two neurologists with at least 5 years of experience. Exclusion criteria included (1) stroke mimics; (2) age >75 years; (3) cerebral haemorrhage, degenerative disease, tumour, or severe hepatic or renal insufficiency; and (4) inability to complete cognitive scales cooperatively because of dysphasia or hearing or visual impairment. The control group (28 patients) included healthy volunteers or patients without TIA/cerebral infarction suffering from dizziness or headaches recruited during the same period; all accepted cranial MRI/CT images and cerebrovascular screenings were used to exclude TIA/MIS. This study was approved by the Ethics Committee of the Qilu Hospital of Shandong University in Qingdao, China. All patients provided written informed consent for participation in this study.
DNA Extraction and Quality Control
Fasting blood samples were drawn using EDTA tubes, and whole-blood DNA was extracted using kits (Tiangen Biotech, Beijing, China). The quality of the DNA was detected by a NanoDrop 2000 (NanoDrop technologies, Wilmington, DE, USA), which required a concentration of ≥20 ng/μL and a total sample purity of ≥400 ng.
CpG Island Selection
CpG islands located in the proximal promoter of the RIN3 gene were selected for measurement according to the following criteria: (1) 200-bp minimum length; (2) 50% or higher GC content; and (3) 0.60 or higher ratio of observed/expected dinucleotide CpG. Two regions from CpG islands of the RIN3 gene were selected and sequenced.
Bisulfite Conversion and Multiplex Amplification
DNA methylation levels were analysed using MethylTarget® (Genesky Biotechnologies Inc., Shanghai, China), an NGS-based multiple targeted CpG methylation analysis method. Specifically, the genomic regions of interest were analysed and transformed into bisulfite-converted sequences using GeneCpG software. Polymerase chain reaction (PCR) primer sets were designed using Methylation Primer software from bisulfate-converted DNA. The PCR primers for RIN3_26 were as follows: forward, 5ʹ-GTATATTTGTTAGGAATGTGGAGGAG-3ʹ; reverse, 5ʹ-AAAAAAAAATCTTCCACTTAACTTAAAACC-3ʹ. The PCR primers for RIN3_27 were as follows: forward, 5ʹ-TTAGTGTTTGGGTAGGGTTTAGG-3ʹ; reverse, 5ʹ-AAACCCAACCCCRAACAA-3ʹ. Genomic DNA (400 ng) was subjected to sodium bisulfite treatment using the EZ DNA Methylation™-GOLD Kit (ZYMO, CA, USA), according to the manufacturer’s protocols. Multiplex PCR was performed using optimised primer set combinations. A 20-µL PCR reaction mixture was prepared for each reaction, which included 10x reaction buffer (TaKaRa, Dalian, China), 25 mM of Mg2+, 2.5 mM of dNTP, 1 µM of each primer, 5U of HotStarTaq polymerase (TaKaRa, Dalian, China), and 1 µL of template DNA. The cycling program included 95°C for 2 min; 11 cycles of 95°C for 20 s, 62°C for 40 s with a decreasing temperature step of 0.5°C per cycle, and 72°C for 1 min; and 24 cycles of 95°C for 20 s, 62°C for 30 s, 72°C for 1 min, and 72°C for 1 min.
Index PCR
PCR amplicons were diluted and amplified using indexed primers. Specifically, a 20-µL mixture was prepared for each reaction comprised of 5x reaction buffer (TaKaRa, Dalian, China), 2.5 mM of dNTP, 10 µM of F primer, 4 µM of index primer, 0.2µL of Herculase® II Fusion DNA polymerase (Agilent Technologies, CA, USA), 2 µL of diluted template, and ddH2O. The cycling program included 95°C for 2 min followed by 11 cycles of 95°C for 20 s, 65°C for 30 s, 72°C for 30 s, and 72°C for 3 min. PCR amplicons (170 bp–270 bp) were separated by agarose electrophoresis and purified using the TIANGEN Gel Extraction kit (TIANGEN, Beijing, China).
Sequencing
Libraries from different samples were quantified and pooled together followed by sequencing on the Illumina HiSeq platform according to the manufacturer’s protocols. Sequencing was performed using a 2 × 150-bp paired-end mode.
Data Analysis
Fast Length Adjustment of SHort reads (FLASH) is an accurate and fast tool used to merge paired-end reads.17 FASTQ to FASTA format was then processed using the Fastx toolkit (http://hannonlab.cshl.edu/fastx_toolkit/index.html). Reads in FASTA format were mapped to the targeted bisulfite genome (hg19) by Blast.18 Unmapped reads were filtered, and mapped reads with a coverage greater than 90% and an identity greater than 90% were considered effective reads and used for the following statistics. The sequencing depth for each amplicon per sample was calculated by blasting the effective reads against the targeted genomic region. Reads less than 10-fold were removed, and the overall sequencing depth for each sample was evaluated. Methylation and haplotypes were analysed using Perl script.
Statistical Methods
R software was used for statistical analysis. Continuous data are presented as mean ± standard deviation (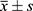 ), and categorical data are expressed as numbers of cases and percentages (n [%]). Student’s t-test was used for continuous variables with normal distribution and homogeneous variance to compare observations between both study groups, and the Wilcoxon test was used otherwise. The test of proportions, including the K-square test and Cochran-Armitage trend test, was applied to determine if there was a statistically significant difference in attribute percentages between data sets. The K-square test was used for nominal categorical variables. The Cochran-Armitage trend test was used for ordinal categorical variables. Single-factor logistic regression and multi-factor logistic regression analyses were performed to identify independent variables for TIS/MIS with early cognitive impairment. We screened variables using a stepwise regression analysis to construct predictive models and plot ROC curves. A p-value <0.05 was considered statistically significant.
), and categorical data are expressed as numbers of cases and percentages (n [%]). Student’s t-test was used for continuous variables with normal distribution and homogeneous variance to compare observations between both study groups, and the Wilcoxon test was used otherwise. The test of proportions, including the K-square test and Cochran-Armitage trend test, was applied to determine if there was a statistically significant difference in attribute percentages between data sets. The K-square test was used for nominal categorical variables. The Cochran-Armitage trend test was used for ordinal categorical variables. Single-factor logistic regression and multi-factor logistic regression analyses were performed to identify independent variables for TIS/MIS with early cognitive impairment. We screened variables using a stepwise regression analysis to construct predictive models and plot ROC curves. A p-value <0.05 was considered statistically significant.
Results
In this study, we tested whole-blood RIN3 methylation in 112 subjects (Figure 1), including 84 patients with TIA/MIS in the case group (group J); 55 patients had early cognitive impairment (group A), whereas 29 participants did not (group B). Moreover, there were 28 participants in the control group (group C). Basic patient clinical information are shown in Tables 1 and 2.
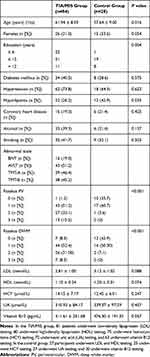 |
Table 1 Clinical Information on the Transient Ischemic Stroke/Mild Ischemic Stroke and Control Groups |
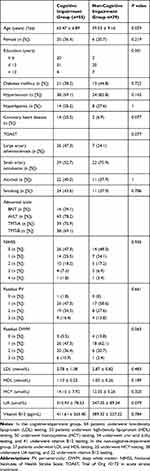 |
Table 2 Clinical Information on the Cognitive-Impairment and Con-Cognitive-Impairment Groups |
Differences Between Patients with TIA/MIS and Healthy Controls in Terms of Total RIN3 Methylation Level
By sequencing methylation in the target regions (Table 3) in the 84 TIA/MIS and 28 controls, we found that the overall methylation level of the RIN3 gene was significantly lower in the TIA/MIS group than in the control group (Groupdiff=−0.02, P <0.001) (Figure 2), even after adjusting for age and gender (Adj. P <0.001) (Table 4).
 |
Table 3 Information on Target DNA Methylation Sequencing |
 |
Table 4 Differences in RIN3 Methylation Levels Between Transient Ischemic Stroke/Mild Ischemic Stroke Patients and Healthy Controls |
Total RIN3 Methylation Levels in Patients with TIA/MIS with and without Cognitive Impairment
The overall methylation level of the RIN3 gene was significantly lower in the cognitive impairment group (n=55) than in the non-cognitive impairment group (n=29) (Groupdiff=−0.013, P=0.01) (Figure 3), even after adjusting for age, gender, education, CHD, and DWM (Adj. P=0.044) (Table 5).
 |
Table 5 Differences in RIN3 Methylation Levels Between Patients with Transient Ischemic Stroke/Mild Ischemic Stroke with and without Cognitive Impairment |
Differences in Differentially Methylated Sites Between Patients with Transient Ischemic Stroke/Mild Ischemic Stroke and Healthy Controls
Whole blood RIN3 gene methylation levels were significantly different between patients with TIA/MIS and controls. We evaluated differences in methylation levels at different sites between both groups and found statistically significant differences at several sites (S26-146, S27-25, 29, 36, 38, 40, 58, 61, 96, 111, 116, 127, 144, 146, and 152, P <0.05), even after adjustment for age and gender. All of these sites showed hypomethylation (Groupdiff <0) in patients with TIA/MIS (Table 6 and Figure 4).
 |
Table 6 Differences in Methylation Sites Between Patients with Transient Ischemic Stroke/Mild Ischemic Stroke and Healthy Controls |
Differences in Differentially Methylated Sites in Patients with Transient Ischemic Stroke/Mild Ischemic Stroke with and without Cognitive Impairment
RIN3 gene methylation levels were significantly different between patients with TIA/MIS with and without cognitive impairment. We examined methylation levels at different sites between both groups and found statistically significant differences (P <0.05) at several sites (S26-113, 165, S27-25, 36, 111, 116, 127, 144, 146, and 152). All of these sites, except site S26-165 (Groupdiff=0.0019, P =0.03), showed hypomethylation in the cognitive impairment group after adjusting for gender, age, education, CHD, and DWM. There groups were not significantly different following adjustment at sites S26-113, 165, S27-36, and 127 (Table 7 and Figure 5).
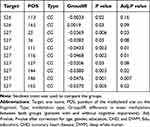 |
Table 7 Different Methylation Sites in Patients with Transient Ischemic Stroke/Mild Ischemic Stroke with and without Cognitive Impairment |
Differences in Differentially Methylated Segments Between Patients with Transient Ischemic Stroke/Mild Ischemic Stroke and Healthy Controls
We investigated whether there were differences in the methylation levels of different fragments between patients with TIA/MIS and healthy controls. By calculating the mean methylation levels of all CpGs of the RIN3 gene, we found that there were statistically significant differences in RIN3-26 (P=0.03) and RIN3-27 (P <0.001) between both groups. The overall methylation levels of RIN3-26 (Groupdiff =−0.0011) and RIN3-27 (Groupdiff =−0.0417) were significantly lower in the TIA/MIS group than in the control group. However, after adjusting for gender and age, there was only a statistically significant difference in RIN3-27 between both groups (Adj. P <0.001) (Table 8).
 |
Table 8 Differences in Differentially Methylated Segments Between Patients with Transient Ischemic Stroke/Mild Ischemic Stroke and Healthy Controls |
Differences in Differentially Methylated Segments Between Patients with TIA/MIS
We investigated whether there were differences in the methylation levels of different fragments between the cognitive impairment and non-cognitive impairment TIA/MIS groups. By calculating the mean methylation levels of all CpGs of the RIN3 gene, we found that there was a statistically significant difference in RIN3-27 between both groups (P=0.01), even after adjusting for age, gender, education, CHD, and DWM (Adj. P=0.046) (Table 9). The methylation level of RIN3-27 was significantly lower (Groupdiff=−0.0268) in the cognitive-impairment group than in the non-cognitive-impairment group.
 |
Table 9 Differences in Differentially Methylated Segments Between Patients with Transient Ischemic Stroke/Mild Ischemic Stroke with and without Cognitive Impairment |
Predictive Model for Acute-Phase TIA/MIS with Cognitive Impairment
After analysing clinical data from the cognitive-impairment and non-cognitive-impairment groups (Table 2) and combining this information with statistically significant differences in RIN3 methylation sites, we constructed a predictive model and plotted ROC curves. We found that the combination of three risk factors, including education level, presence of coronary heart disease, and S27-146 site methylation, showed the highest sensitivity/specificity for predicting early cognitive impairment after TIA/MIS. Thus, this model was able to scientifically forecast the occurrence of early cognitive impairment after TIA/MIS, with an area under the curve of 0.808 (95% CI: 0.7119–0.9044) (Figure 6).
 |
Figure 6 Showed the sensitivity/specificity for predicting early cognitive impairment after TIA/MIS. |
Discussion
This study demonstrated that hypomethylation of the RIN3 gene was associated with TIA/MIS and with early cognitive impairment after its onset. Furthermore, a combination of clinical indicators and methylation sites could predict the onset of early cognitive impairment after TIA/MIS. At present, there are few studies related to the relationship between gene methylation and early cognitive impairment after TIA/MIS, prompting this study.
Currently, there is no standardised scale for measuring vascular cognitive impairment. In this study, we used four scales to detect cognitive impairment in individual domains and found that the incidence of cognitive impairment after TIA/MIS was 65.5% (55/84), which is consistent with results reported in other studies.1,3,4 Thus, the results obtained from these scales reflected the cognitive status of patients at that time. We investigated the correlation between RIN3 methylation and TIA/MIS and between RIN3 methylation and TIA/MIS with early cognitive impairment. Our results showed that the RIN3 gene was hypomethylated in the whole blood of patients with TIA/MIS relative to a healthy control group. In addition, there was hypomethylation of the RIN3 gene in TIA/MIS patients with acute-phase cognitive impairment in comparison to TIA/MIS patients without acute-phase cognitive impairment. Shen et al have shown that increased expression levels of RIN3 can affect its transport function, which may lead to an increase in the αβ protein within neurons. Boden found that patients with early-onset AD showed hypomethylation of the RIN3 gene in whole blood, which led to higher expression of the RIN3 gene.8 Another study confirmed that αβ plays a role in vascular cognitive impairment.19 Therefore, we hypothesized that patients with TIA/MIS would also show abnormal methylation of the RIN3 gene.
Cognitive impairment is common within 7 days after onset of TIA/MIS and may persist even after recovery of motor symptoms, leading to a greater risk of cognitive impairment and a reduced cognitive reserve in the distant future. Despite the rapid recovery of motor symptoms in TIA/MIS patients, cognitive impairment can easily be overlooked in these patients.2 The occurrence of early cognitive impairment after TIA/MIS is influenced by multiple factors, and the current study found that some imaging changes may serve as predictors. Suda et al found that temporal horn atrophy on MRI education and smoking are independent risk factors for cognitive impairment.1 Takahashi also confirmed that moderate temporal lobe atrophy on MRA source images combined with less years of education were predictors of cognitive impairment in the acute phase of ischemic stroke.20 Finally, Zamboni found that white-matter brain damage on MRI was associated with TIA/MIS with early cognitive impairment damage.21 These factors are, to some extent, nonmodifiable. Furthermore, it can be difficult to standardise their quantitative detection criteria. To date, there has been a lack of identification of modifiable biomolecular indicators related to disease onset, as well as a lack of quantifiable blood molecular indicators. This study, however, found that the RIN3 gene and abnormal methylation of sites on this gene are associated with early cognitive impairment after TIA/MIS. Since methylation levels can be affected by environmental factors and can be measured quantitatively, they may be useful for the design of potential interventions and predictive models.
There is an increasing number of studies related to the correlation between methylation and ischemic stroke.22–28 For example, methylation of the ABCG1 and APOE genes has been correlated with cerebral infarction and atherosclerosis,23 while MMP-2, LINE3 and TP53 methylation has been correlated with ischemic stroke,24–26 and CDKN2B has been correlated with arterial calcification in ischemic stroke.27,28 Furthermore, methylation has been strongly correlated with the pathogenesis of ischemic stroke. However, these studies have mainly focused on the relationship between gene methylation and ischemic stroke or atherosclerosis rather than specifically focusing on TIA/MIS. Moreover, there have been few studies related to the relationship between methylation and early cognitive impairment after stroke.
Some studies have investigated molecular indicators in the cerebrospinal fluid of patients with vascular dementia (VD) or VCI and have found that Aβ42 are significantly decreased in the cerebrospinal fluid of patients with VCI compared to controls.29–33 Aβ concentration in red blood cell of VD patients is higher than that of controls.34 Elevated Aβ levels in blood may aggravate vascular amyloidosis and thus affect cognition.35 However, whether Aβ in blood can predict VCI still requires verification. NF-κB and VEGF levels have also been shown to be elevated in the cerebrospinal fluid of patients with VD, which may affect the β-amyloid protein.36,37 Abnormal expression of the Aβ protein has been proposed to be significantly associated with vascular-related cognitive impairment.
The RIN3 gene located on chromosome 14, however, has been less well studied. This gene has, however, been found to affect endocytosis of axonemes by interacting with BIN1, which influences the transport and metabolism of the Aβ protein, with high expression of the RIN3 gene decreasing Aβ protein metabolism.5,6 It has also been shown that abnormal RIN3 gene expression is associated with AD7 and that whole-blood RIN3 methylation levels are low in AD patients.8 RIN3 affects cognitive function mainly by influencing Aβ protein transport. It is therefore likely that the appearance of early cognitive impairment in TIA/MIS patients is also associated with abnormal RIN3 gene expression and with altered Aβ protein levels. The methylation level is an important factor affecting gene expression. This study confirmed that abnormal RIN3 gene methylation was associated with early cognitive impairment in TIA/MIS.
This study examined RIN3 methylation levels in whole blood and found that the RIN3 gene was hypomethylated in the TIA/MIS group compared to the control group and that RIN3 methylation levels were lower in TIA/MIS patients with early cognitive impairment than in those without cognitive impairment. We hypothesize that RIN3 hypomethylation occurs during the acute phase of TIA and mild stroke, which, in turn, leads to an abnormal accumulation of the Aβ protein in the blood. Indeed, this study did find a correlation between abnormal RIN3 methylation in blood and TIA/MIS with early cognitive impairment; however, the causal relationship between the two remains unclear. Based on our study, it is not possible to determine whether RIN3 gene hypomethylation in the blood led to an accumulation of the Aβ protein, which resulted in the appearance or exacerbation of symptoms, or whether the disease itself first affected the level of RIN3 methylation, which in turn affected the Aβ protein level in blood. These issues require further research.
This study had some limitations. First, the number of cases in this study was small and not fully matched by age nor sex. However, we corrected for age and sex to minimise the influence of these factors on our statistical analyses. Second, this study was designed to explore biomarkers in whole blood, so we examined DNA methylation in whole blood. However, we cannot confirm whether methylation levels in whole blood are reflected in methylation levels in brain tissue. However, we chose to test whole blood because the underlying cause of cerebrovascular disease involves changes to the blood vessels and the blood itself, which, in turn, leads to brain-tissue damage.
Conclusion
This study confirmed that there was hypomethylation of the RIN3 gene in the whole blood of TIA/MIS patients relative to controls. Furthermore, there was hypomethylation of the RIN3 gene in the whole blood of patients with early cognitive impairment after TIA/MIS relative to patients without early cognitive impairment after TIA/MIS. Therefore, epigenetic modification of the RIN3 gene was strongly associated with TIA/MIS and with TIA/MIS with early cognitive impairment. Based on the modifiable nature of methylation, our study suggests that it may be possible to influence this disease process by methylation via appropriate lifestyle and/or clinical interventions. Furthermore, since methylation can be quantified and easily analysed, the methylation levels of gene sites combined with clinical information can predict the occurrence of TIA/MIS with early cognitive impairment.
Data Sharing Statement
Datasets and codes of methylation used in the analyses are stored by the first author and will be provided upon request.
Ethics
This study was approved by the Ethics Committee of the Qilu Hospital of Shandong University (Qingdao). All patients provided written informed consent. The study complies with the Declaration of Helsinki.
Acknowledgments
This work was supported by Qingdao Key Health Discipline Development Fund and Scientific Research Foundation of Qilu Hospital of Shandong University (Qingdao) (No.QDKY2019ZD02)
Author Contributions
All authors made a significant contribution to the work reported, whether that is in the conception, study design, execution, acquisition of data, analysis and interpretation, or in all these areas; took part in drafting, revising or critically reviewing the article; gave final approval of the version to be published; have agreed on the journal to which the article has been submitted; and agree to be accountable for all aspects of the work.
Disclosure
The authors declare that there is no conflicts of interest or financial relationships that could be construed as a potential conflict of interest in this work.
References
1. Suda S, Nishimura T, Ishiwata A, et al. Early cognitive impairment after minor stroke: associated factors and functional outcome. J Stroke Cerebrovasc Dis. 2020;29(5):104749. doi:10.1016/j.jstrokecerebrovasdis.2020.104749
2. Pendlebury ST, Wadling S, Silver LE, Mehta Z, Rothwell PM. Transient cognitive impairment in TIA and minor stroke. Stroke. 2011;42(11):3116–3121. doi:10.1161/STROKEAHA.111.621490
3. Blackburn DJ, Bafadhel L, Randall M, Harkness KA. Cognitive screening in the acute stroke setting. Age Ageing. 2013;42(1):113–116. doi:10.1093/ageing/afs116
4. Godefroy O, Fickl A, Roussel M, et al. Is the montreal cognitive assessment superior to the mini-mental state examination to detect poststroke cognitive impairment? A study with neuropsychological evaluation. Stroke. 2011;42(6):1712–1716. doi:10.1161/STROKEAHA.110.606277
5. Kajiho H, Saito K, Tsujita K, et al. RIN3: a novel Rab5 GEF interacting with amphiphysin II involved in the early endocytic pathway. J Cell Sci. 2003;116(Pt20):4159–4168. doi:10.1242/jcs.00718
6. Kajiho H, Sakurai K, Minoda T, et al. Characterization of RIN3 as a guanine nucleotide exchange factor for the Rab5 subfamily GTPase Rab31. J Biol Chem. 2011;286(27):24364–24373. doi:10.1074/jbc.M110.172445
7. Shen R, Zhao X, He L, et al. Upregulation of RIN3 induces endosomal dysfunction in Alzheimer’s disease. Transl Neurodegener. 2020;9(1):26. doi:10.1186/s40035-020-00206-1
8. Boden KA, Barber IS, Clement N, et al. Methylation profiling RIN3 and MEF2C identifies epigenetic marks associated with sporadic early onset alzheimer’s disease. J Alzheimers Dis Rep. 2017;1(1):97–108. doi:10.3233/ADR-170015
9. Tian JZ, Xie HG, Qin B, et al. Screening and diagnostic framework of vascular dementia in Chinese population. Zhonghua Nei Ke Za Zhi. 2019;58(1):10–16. doi:10.3760/cma.j.issn.0578-1426.2019.01.003
10. Hao L, Xing Y, Li X, et al. Risk factors and neuropsychological assessments of subjective cognitive decline (plus) in chinese memory clinic. Front Neurosci. 2019;13:846. doi:10.3389/fnins.2019.00846
11. Ma J, Zhang Y, Guo Q. Comparison of vascular cognitive impairment–no dementia by multiple classification methods. Int J Neurosci. 2015;125(11):823–830. doi:10.3109/00207454.2014.972504
12. Xu Y, Chen K, Zhao Q, Li F, Guo Q. Short-term delayed recall of auditory verbal learning test provides equivalent value to long-term delayed recall in predicting MCI clinical outcomes: a longitudinal follow-up study. Appl Neuropsychol Adult. 2020;27(1):73–81. doi:10.1080/23279095.2018.1481067(AVLT
13. Llinàs-Reglà J, Vilalta-Franch J, López-Pousa S, Calvó-Perxas L, Torrents Rodas D, Garre-Olmo J. The trail making test. Assessment. 2017;24(2):183–196. doi:10.1177/1073191115602552
14. Sánchez-Cubillo I, Periáñez JA, Adrover-Roig D, et al. Construct validity of the trail making test: role of task-switching, working memory, inhibition/interference control, and visuomotor abilities. J Int Neuropsychol Soc. 2009;15(3):438–450. doi:10.1017/S1355617709090626
15. Wei M, Shi J, Li T, et al. Diagnostic accuracy of the chinese version of the trail-making test for screening cognitive impairment. J Am Geriatr Soc. 2018;66(1):92–99. doi:10.1111/jgs.15135
16. Gorelick PB, Counts SE, Nyenhuis D. Vascular cognitive impairment and dementia. Biochim Biophys Acta. 2016;1862(5):860–868. doi:10.1016/j.bbadis.2015.12.015
17. Magoč T, Salzberg SL. FLASH: fast length adjustment of short reads to improve genome assemblies. Bioinformatics. 2011;27(21):2957–2963. doi:10.1093/bioinformatics/btr507
18. Camacho C, Coulouris G, Avagyan V, et al. BLAST+: architecture and applications. BMC Bioinform. 2009;10:421. doi:10.1186/1471-2105-10-421
19. Cipollini V, Troili F, Giubilei F. Emerging biomarkers in vascular cognitive impairment and dementia: from pathophysiological pathways to clinical application. Int J Mol Sci. 2019;20(11):2812. doi:10.3390/ijms20112812
20. Takahashi Y, Saito S, Yamamoto Y, et al. Visually-rated medial temporal lobe atrophy with lower educational history as a quick indicator of amnestic cognitive impairment after stroke. J Alzheimers Dis. 2019;67(2):621–629. doi:10.3233/JAD-180976
21. Zamboni G, Griffanti L, Jenkinson M, et al. White matter imaging correlates of early cognitive impairment detected by the montreal cognitive assessment after transient ischemic attack and minor stroke. Stroke. 2017;48(6):1539–1547. doi:10.1161/STROKEAHA.116.016044
22. Deng GX, Xu N, Huang Q, et al. Association between promoter DNA methylation and gene expression in the pathogenesis of ischemic stroke. Aging (Albany NY). 2019;11(18):7663–7677. doi:10.18632/aging.102278
23. Qin X, Li J, Wu T, et al. Overall and sex-specific associations between methylation of the ABCG1 and APOE genes and ischemic stroke or other atherosclerosis-related traits in a sibling study of Chinese population. Clin Epigenetics. 2019;11(1):189. doi:10.1186/s13148-019-0784-0
24. Lin HF, Hsi E, Huang LC, et al. Methylation in the matrix metalloproteinase-2 gene is associated with cerebral ischemic stroke. J Investig Med. 2017;65(4):794–799. doi:10.1136/jim-2016-000277
25. Wei Y, Sun Z, Wang Y, et al. Methylation in the TP53 promoter is associated with ischemic stroke. Mol Med Rep. 2019;20(2):1404–1410. doi:10.3892/mmr.2019.10348
26. Lin RT, Hsi E, Lin HF, Liao YC, Wang YS, Juo SH. LINE-1 methylation is associated with an increased risk of ischemic stroke in men. Curr Neurovasc Res. 2014;11(1):4–9. doi:10.2174/1567202610666131202145530
27. Zhou S, Zhang Y, Wang L, et al. CDKN2B methylation is associated with carotid artery calcification in ischemic stroke patients. J Transl Med. 2016;14:333. doi:10.1186/s12967-016-1093-4
28. Zhou S, Cai B, Zhang Z, et al. CDKN2B methylation and aortic arch calcification in patients with ischemic stroke. J Atheroscler Thromb. 2017;24(6):609–620. doi:10.5551/jat.36897
29. Llorens F, Schmitz M, Knipper T, et al. Cerebrospinal fluid biomarkers of alzheimer’s disease show different but partially overlapping profile compared to vascular dementia. Front Aging Neurosci. 2017;9:289. doi:10.3389/fnagi.2017.00289
30. Ewers M, Mattsson N, Minthon L, et al. CSF biomarkers for the differential diagnosis of Alzheimer’s disease: a large-scale international multicenter study. Alzheimers Dement. 2015;11(11):1306–1315. doi:10.1016/j.jalz.2014.12.006
31. Struyfs H, Van Broeck B, Timmers M, et al. Diagnostic accuracy of cerebrospinal fluid amyloid-β isoforms for early and differential dementia diagnosis. J Alzheimers Dis. 2015;45(3):813–822. doi:10.3233/JAD-141986
32. Duits FH, Hernandez-Guillamon M, Montaner J, et al. Matrix metalloproteinases in alzheimer’s disease and concurrent cerebral microbleeds. J Alzheimers Dis. 2015;48(3):711–720. doi:10.3233/JAD-143186
33. Horvath I, Jia X, Johansson P, et al. Pro-inflammatory S100A9 protein as a robust biomarker differentiating early stages of cognitive impairment in alzheimer’s disease. ACS Chem Neurosci. 2016;7(1):34–39. doi:10.1021/acschemneuro.5b00265
34. Lauriola M, Paroni G, Ciccone F, et al. Erythrocyte associated amyloid-β as potential biomarker to diagnose dementia. Curr Alzheimer Res. 2018;15(4):381–385. doi:10.2174/1567205014666171110160556
35. Yamada M. Cerebral amyloid angiopathy: emerging concepts. J Stroke. 2015;17(1):17–30. doi:10.5853/jos.2015.17.1.17
36. Buchhave P, Zetterberg H, Blennow K, Minthon L, Janciauskiene S, Hansson O. Soluble TNF receptors are associated with Aβ metabolism and conversion to dementia in subjects with mild cognitive impairment. Neurobiol Aging. 2010;31(11):1877–1884. doi:10.1016/j.neurobiolaging.2008.10.012
37. Tarkowski E, Issa R, Sjögren M, et al. Increased intrathecal levels of the angiogenic factors VEGF and TGF-beta in Alzheimer’s disease and vascular dementia. Neurobiol Aging. 2002;23(2):237–243. doi:10.1016/s0197-4580(01)00285-8
 © 2021 The Author(s). This work is published and licensed by Dove Medical Press Limited. The full terms of this license are available at https://www.dovepress.com/terms.php and incorporate the Creative Commons Attribution - Non Commercial (unported, v3.0) License.
By accessing the work you hereby accept the Terms. Non-commercial uses of the work are permitted without any further permission from Dove Medical Press Limited, provided the work is properly attributed. For permission for commercial use of this work, please see paragraphs 4.2 and 5 of our Terms.
© 2021 The Author(s). This work is published and licensed by Dove Medical Press Limited. The full terms of this license are available at https://www.dovepress.com/terms.php and incorporate the Creative Commons Attribution - Non Commercial (unported, v3.0) License.
By accessing the work you hereby accept the Terms. Non-commercial uses of the work are permitted without any further permission from Dove Medical Press Limited, provided the work is properly attributed. For permission for commercial use of this work, please see paragraphs 4.2 and 5 of our Terms.





