Back to Journals » Journal of Inflammation Research » Volume 14
Methyl Diet Enhanced Sepsis-Induced Mortality Through Altering Gut Microbiota
Authors Yu C, Zhu X, Zheng C, Luo Y, Wang F, Gao Y, Wu H, Sun X, Kong X
Received 4 February 2021
Accepted for publication 29 June 2021
Published 9 July 2021 Volume 2021:14 Pages 3107—3121
DOI https://doi.org/10.2147/JIR.S305202
Checked for plagiarism Yes
Review by Single anonymous peer review
Peer reviewer comments 2
Editor who approved publication: Dr Monika Sharma
Chang Yu,1,* Xiaojun Zhu,1,* Chao Zheng,1 Yichun Luo,1 Fang Wang,1 Yueqiu Gao,1 Hailong Wu,2 Xuehua Sun,1 Xiaoni Kong1
1Central Laboratory, Department of Liver Diseases, ShuGuang Hospital Affiliated to Shanghai University of Chinese Traditional Medicine, Shanghai, People’s Republic of China; 2Shanghai Key Laboratory of Molecular Imaging, Shanghai University of Medicine and Health Sciences, Shanghai, People’s Republic of China
*These authors contributed equally to this work
Correspondence: Xiaoni Kong; Xuehua Sun
Central Laboratory, Department of Liver Diseases, ShuGuang Hospital Affiliated to Shanghai University of Chinese Traditional Medicine, 528 Zhangheng Road, Shanghai, 201203, People’s Republic of China
Tel +86 21 20256838
Email [email protected]; [email protected]
Introduction: Mortality of sepsis is caused by an inappropriately amplified systemic inflammatory response and bacteremia. Methyl diet has been shown to associate with greater inflammation response in different diseases. This study aimed to determine whether dietary supplementation with methyl donors affects the inflammation response and mortality in sepsis and to investigate the underlying mechanisms.
Methods: Four-week-old male C57BL/6 mice were fed with a high-methyl diet (HMD) or a regulator diet (RD) till the experiment time. Mice septic model was induced by Cecal ligation and puncture (CLP), lipopolysaccharide (LPS), or E.coli. Inflammatory cytokine was analyzed by ELISA and qRT-PCR. Immune cell infiltration was evaluated by H&E and IHC. The composition of gut microbiota was determined by 16S rRNA sequencing. The effect of gut microbiota on sepsis was further verified by fecal microbiome transplantation.
Results: Our results showed that the diet riches in methyl donors exacerbated mortality, organ injury, and circulating levels of inflammatory mediators in CLP-induced septic mice model, compared to the control diet group. However, no significant differences have been observed in the inflammatory responses in the LPS-induced septic model and macrophages activation between the two groups of mice. There was a higher bacterial burden in CLP-induced HMD mice suggested that methyl diet might modulate gut microbiota. Bacterial 16S rRNA sequencing results showed that the composition of gut microbiota was altered. The high methyl donor diet reduced the abundance of Akkermansia and Lachnospiraceae, which were associated with protective effects in sepsis, in the gut. Moreover, fecal microbiome transplantation experiment showed that the transfer of feces, which obtained from high methyl diet mice, aggravated the mortality and inflammation responses in recipient mice.
Discussion: Methyl diet enhanced CLP-induced septic mortality and inflammatory responses through altering the composition of gut microbiota. This result indicated that diet-based gut microbiota may be a new therapeutic strategy for sepsis patients.
Keywords: sepsis, DNA methylation, gut microbiota, Akkermansia, Lachnospiraceae
Introduction
Sepsis is defined as life-threatening organ dysfunction caused by a dysregulated host response to infection.1 Due to its high mortality rate, sepsis is still regarded as a global health problem.2,3 Sepsis is an intrinsic heterogeneity, multifactorial disease influenced by pathogenic factors, host factors, and environmental factors.4–7 For decades, studies have shown that gut functionality and microbial diversity play important roles in sepsis pathogenesis.8–10 In the homeostatic condition, the gut microbiota protects the host against systemic infection by inducing the generation of IgG, IgA, and antimicrobial peptides.10–12 At the same time, the microbiota that is derived from the gut is a major cause of multiple organ dysfunction in sepsis.13,14 Clinical studies also proved that the gut microbiota had been disturbed in sepsis patients.15
Lifestyle is a crucial factor that shapes the gut microbiota, including diet, antibiotics, aging, and etc.15–17 Studies across mice and humans suggest that common aspects of the lifestyle, including the high-fat diet, can persistently alter commensal microbial communities.18 Chassaing and colleagues showed that dietary emulsifiers can promote colitis and metabolic syndrome by affecting the mouse gut microbiota.16 Mice were fed with non-fermentable fiber impact the composition of gut microbiota improving sepsis.6
Methyl-related nutrients are found in three groups of compounds: B Vitamins, Betaine, and S-Adenosyl-Methionine (SAMe). Methyl-related nutrients can be found in foods that are rich in folate, which is the natural form of Vitamin B9, or choline, which can be metabolized into betaine. Folic acid and betaine are the important methyl donors in the DNA methylation process.19–21 Cyanocobalamin is also known as vitamin B12, which works as a cofactor with methionine synthase (MS) to catalyze the renewal of methionine.19 The effects of methyl-related nutrients on health are controversial. There are few studies about the effects of the methyl diet on intestinal flora and sepsis.
In our study, we fed mice (4-weeks old just after weaning time) with high-methyl diet (HMD), which is supplemented with methyl donors and cofactors (such as folic acid, betaine, cyanocobalamin, choline, and etc). By using the cecal ligation and puncture (CLP) induced sepsis model, we found that, compared to the normal diet group of mice, HMD group mice showed a higher mortality rate and greater systemic inflammation responses. However, there was no significant difference in the LPS and E.coli-induced septic models. Mechanistically, we found that the diet riches in methyl donors reduced the abundance of Akkermansia and Lachnospiraceae, which were associated with metabolic health protective effects in sepsis, in the gut. Moreover, fecal microbiome transplantation experiment showed that the transfer of feces, which obtained from HMD group mice, aggravated the disease mortality and inflammation response in recipient mice. This further supports that the high-methyl diet enhances sepsis mortality by altering the gut microbiota.
Materials and Methods
Methyl Diet and Feeding Protocol
Four-week-old male C57BL/6 mice (Shanghai Model Organisms, Shanghai, China) were fed with a high-methyl diet (HMD) or a regulator diet (RD) till the experiment time. Four-week-old male SD rats (Shanghai Model Organisms, Shanghai, China) were fed with a HMD or a RD for eight weeks. Two kinds of diets were purchased from the Research Diets company (New Brunswick, NJ, United States). HMD includes folic acid, betaine, cyanocobalamin, choline, etc. Folic acid and betaine are the major methyl donors, and cyanocobalamin works as a cofactor to catalyze methionine renew.19 The detailed formulation of diets (HMD and RD) is listed in Table 1. All procedures involving animals were approved and performed in accordance with the Animal Care and Use Committee of Shanghai University of Chinese Traditional Medicine.
 |
Table 1 The Formulation of Diets |
Septic Mice Models
The cecal ligation and puncture (CLP) induced septic model was described previously.7,22,23 Briefly, mice were anesthetized with isoflurane/oxygen inhalational gas (Shanghai Yuyan, Shanghai, China). The skin midline of the abdomen was incised and then the cecum was ligated at different positions for the desired severity grade (survival: 1/2 distance from ileocecal valve; septic model: 2/3 distance from ileocecal valve). Then, the cecum was punctured, a few cecal contents were pushed and the cecum was relocated back into the peritoneal cavity. Finally, the peritoneum was closed. In the LPS-induced septic model, 5mg/kg (sublethal dose) or 10mg/kg (lethal dose) LPS (Sigma, St. Louis, MO, United States) was intraperitoneally injected into mice.
Serum and Peritoneal Lavage Fluid Cytokines Analysis
The levels of IL-6 and TNF-α were detected by enzyme-linked immunosorbent assay (ELISA) kits (NeoBioscience, Shenzhen, Guangdong, China) according to the manufacturer’s instructions.
Isolation of Peritoneal Macrophages
Peritoneal macrophages were isolated as described.24 Macrophages were cultured in RPMI 1640 medium with 10% fetal bovine serum with 1% antibiotics (Gibco, Co Dublin, Ireland) overnight. The medium was instated with RPMI 1640 medium for 1h. And then cells were treated with 100ng/mL LPS. Cells were harvested 4 hours later for IL-6 and TNF-α Real-time PCR measurement, and the supernatant was collected 6 hours later for IL-6 and TNF-α Elisa measurement. Primer for IL-6, forward: 5′-TGTTCTCTGGGAAATCGTGGA-3′; reverse: 5′-TTTCTGCAAGTGCATCATCGT-3′. Primer for TNF-α, forward: 5′-TTCTATGGCCCAGACCCTCA-3′; reverse: 5′-TTTGCTACGACGTGGGCTAC-3′. GAPDH was used as internal control, forward: 5′-AGGCCGGTGCTGAGTATGTC-3′; reverse: 5′-TGCCTGCTTCACCACCTTCT-3′. Akkermansia, forward: 5′-CAGCACGTGAAGGTGGGGAC-3′; reverse 5′-CCTTGCGGTTGGCTTCAGAT-3′.25
Stool DNA Extraction
Feces were firstly collected in tubes and the corresponding weights were recorded. DNA was extracted from two groups of rats’ stool samples using QIAamp Fast DNA Stool Mini Kit (QIAGEN, Hilden, Germany) according to the manufacturer’s instructions. Real-time PCR was used to analyze the concentration of Akkermansia. Standard curves were created using serial 10-fold dilutions of E.coli. The bacterial concentration of each sample was calculated according to the standard curve.
Tissue Histopathology, Immunohistochemical Staining
Liver and lung tissues were fixed in 4% paraformaldehyde (Beijing Dingguo Changsheng Biotechnology, Beijing, China), then embedded in paraffin. Then, 5-μm sections were cut and used to stain Hematoxylin and eosin (Beyotime, Shanghai, China) or Myeloperoxidase (MPO, Biocare Medical, Pacheco, CA, United States) antibody for immunohistochemical staining as previously described.26
Fecal Microbiota 16S rRNA Sequencing
Stool DNA was extracted by QIAamp fast DNA stool mini kit (QIAGEN, Hilden, Germany) according to the manufacturer’s protocol and DNA quality was checked by agarose gel electrophoresis. 16S rRNA sequencing was conducted by Majorbio (Shanghai, China), 16S rRNA gene, V3-V4 hypervariable regions, were amplified with primers 338F (5′- ACTCCTACGGGAGGCAGCAG-3′) and 806R (5′-GGACTACHVGGGTWTCTAAT-3′) by thermocycler PCR system (GeneAmp 9700, ABI, Waltham, MA, United States). Purified amplicons were pooled in equimolar and paired-end sequenced on an Illumina MiSeq platform (Illumina, San Diego, CA, United States) according to the standard protocols of Majorbio Bio-Pharm Technology Co. Ltd. (Shanghai, China). Raw fast files were quality-filtered by Trimmomatic and merged by FLASH. Operational taxonomic units (OTUs) were clustered with 97% similarity cutoff using UPARSE (version 7.1 http://drive5.com/uparse/) with a novel “greedy” algorithm that performs chimera filtering and OTU clustering simultaneously. The taxonomy of each 16S rRNA gene sequence was analyzed by the RDP Classifier algorithm (http://rdp.cme.msu.edu/) against the Silva (SSU123) 16S rRNA database using a confidence threshold of 70%. These data were analyzed on the free online platform of Majorbio Cloud Platform (www.majorbio.com).
Fecal Microbiome Transplantation Experiment
Mice were gavaged with the mixture of antibiotics (ampicillin, neomycin (Cayman, Ann Arbor, MI, United States), metronidazole (Sigma, St. Louis, MO, United States) 1mg/L; vancomycin (Cayman, Ann Arbor, MI, United States) (0.5mg/mL) three consecutive days to deplete the gut microbiota. On the fourth day after antibiotics treatment, mice were orally administered 200μL fecal suspension, which was retrieved from RD and HMD mice and was suspended in 30% glycerol (Sangon Biotech, Shanghai, China) diluted in PBS (Gibco, Co Dublin, Ireland) (100mg/1.2mL), for totally four times. Fecal microbiome transplanted mice were used for experiments.
Statistical Analysis
Statistical analysis was performed using GraphPad Prism software 7.0. Data are expressed as mean ± SEM. Unpaired Student’s t-test was used to compare two groups. Survival curves were compared using the log-rank (Mantel-Cox) test. 16S rRNA sequencing data were analyzed on the free online platform of Majorbio Cloud Platform (www.majorbio.com).
Results
Methyl Diets Enhanced Systemic Inflammation and Mortality in the CLP-Induced Septic Model
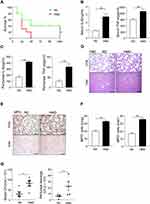
Mice were feed with the regular diet (RD) or a diet rich in methyl donor (HMD) (Table 1) at the time of after weaning. The weight of mice was recorded every week. The result showed that the weight of mice in the HMD group was lower than that of mice in the RD group (Figure S1). Four weeks later, all mice from both groups were subjected to CLP-induced septic model and survival rates were observed. The mortality rates in the HMD group of mice were up to 70% at 24h and reached 100% at 48h after CLP (Figure 1A). However, mortality rates of the RD group of mice were significantly reduced (60% at 48h) and 40% of mice in the RD group were still survived after 5 days (Figure 1A). We then investigated the effects of HMD on inflammation responses in a sublethal CLP model, and there were significantly higher serum and peritoneal levels of proinflammatory cytokines (IL-6, TNF-α) in the HMD group compared to the RD group (Figure 1B and C). Consistent with the higher systemic inflammation, there were also higher number of total immune cells and neutrophils in lung (Figure 1D–F) in the HMD group compared with the RD ones, which suggests that there were higher systemic and local inflammatory responses that caused higher mortality. These results indicate that dietary supplementation with methyl donors enhanced CLP-induced sepsis mortality and inflammation response.
Methyl Diet Does Not Significantly Increase the Mortality and Inflammation in the LPS-Induced Septic Model
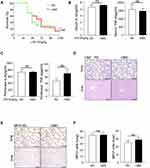
LPS is the major component of the outer membrane of Gram-negative bacteria and it can induce sepsis by inducing an inappropriate amplification of the immune response,27 so it is commonly used to induce sepsis model. Therefore, we investigated the mortality and inflammation response by using the LPS model. Surprisingly, we found that, contrary to the CLP-induced sepsis model, there was no significant difference in the mortality rate between the HMD and the RD groups of mice (Figure 2A). At the same time, there was also no difference in the release of pro-inflammatory cytokines and the recruited immune cells in both groups of mice (Figure 2B–F). Moreover, we also adopted E.coli induced peritonitis model. Mice were treated with the 1×107 E.coli. After 24h, we found the level of inflammatory cytokines and the infiltration of immune cells had no significant difference between RD and HMD mice (Figure 3A–F), which is consistent with the result was the same as that of mice treated with LPS. Thus, the differences in the CLP and the LPS models suggested that the response of innate immune cells might not be the main contribution to the high mortality in the CLP model.
LPS-Stimulated Macrophages Activation Showed No Significant Differences from HMD and RD Fed Mice
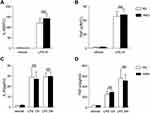
To further analyze whether the innate immune cell responses are different in the HMD and the RD group, we then examined the responses of LPS-treated macrophages from HMD- or RD-fed mice. Compared to the macrophages from the RD group, both in mRNA and protein levels of IL-6 and TNF-α from the HMD group macrophages showed no significant differences (Figure 4A–D). Although the previous study has shown that DNA methylation might modify immune responses during infection,28 in our model, the activation of macrophages between HMD and RD was not significantly different and, more importantly, there were no differences in LPS or E.coli induced mice model (Figures 2–3). This suggested that the difference in the activation of macrophages was not the main reason contributing to the high mortality in the CLP model of HMD-fed mice. We then determined whether the bacterial burdens were contributed to the higher mortality of the CLP model in the HMD group. As shown in Figure 1G, there was a significantly higher bacterial burden in both blood and peritoneal cavity of HMD groups of mice compared with the RD mice. These results show that the higher mortality of HMD mice from CLP-induced sepsis is caused by increased bacterial burden.
Methyl Supplementation Changes the Gut Microbiota Composition
Gut microbiota has been shown to play an important role in sepsis pathogenesis8 and gut microbiota is the infection source in the CLP model, which suggests that HMD may change the microbiota composition. In order to verify whether microbiota composition was altered in HMD mice, we investigated the microbiota composition by 16S rRNA sequencing. As shown in Figure 5A, the amount of sequencing data is reasonable. We found that there was no significant difference in α-diversities (Ace, Chao, Shannon indicated that the species richness and diversity of microbiota) between mice from two groups (Figure 5B). Further hierarchical clustering analysis, including Principal component analysis (PCA) and Principal co-ordinates analysis (PCoA), found distinctly different gut microbiota profiles in the RD and the HMD groups of mice (Figure 5C). HMD mice had a higher number of Firmicutes and a lower number of Verrucomicrobia than RD mice in phylum-level, which suggests that administration of methylation diet destabilized and altered microbiota composition (Figure 5D). Meanwhile, at the genus level, the number of Lachnoclostridium, Blautia was increased and the number of Akkermansia, Lachnospiraceae_NK4A136_group was reduced in HMD mice (Figure 5E). Both bacteria are associated with metabolic health protective effects in sepsis.29 Thus, the methyl diet altered the gut microbiota composition and decreased helpful bacteria for sepsis.
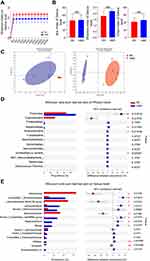 |
Figure 5 The composition of gut microbiota is altered by methyl donor supplement. Fecal samples from RD and HMD groups of mice were analyzed by 16S rRNA sequencing. Dilution curves (A), α-diversity (B), β-diversity (C), and species composition (D, E) were in A-E. The data were analyzed on the free online platform of Majorbio Cloud Platform (www.majorbio.com). The data are shown as means ± SD. *P < 0.05, **P < 0.01. Abbreviation: NS, no significant difference. |
Fecal from HMD Mice Increased the Inflammation and Mortality in CLP-Induced Sepsis in RD Mice
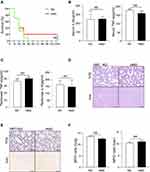
The gut microbiome has been shown to be related to many diseases, including inflammatory bowel disease (IBD),30–32 experimental autoimmune encephalomyelitis (EAE),33 gastric cancer,34 obesity,35 and diabetes.36 We have found that the diet riches in methyl donors could alter the gut microbiota composition. To explore whether the higher septic mortality and inflammation in HMD mice are caused by the alteration in microbiota, we adopted the Fecal microbiome transplantation (FMT) experiment. After antibiotics treatment to deplete the gut microbiota, the microbiota of RD mice was transferred into RD (RD-RD) and HMD mice (RD-HMD) mice or the microbiota of HMD mice was transferred into HMD mice (HMD-HMD) and RD mice (HMD-RD). We found that the mice transplanted with feces from the RD groups showed a higher survival rate than the mice received feces from the HMD groups (Figure 6A). Meanwhile, RD-HMD and RD-RD group had a lower level of pro-inflammatory cytokines (IL-6, TNF-α) than HMD-RD group and HMD-HMD group (Figure 6B and C). Also, there was more neutrophils infiltration in mice that received microbiota from HMD mice than RD mice (Figure 6D and E). In addition, we also measured the change of intestinal flora after methylated diet in rats CLP model. Four-week-old male SD rats were fed with HMD or RD. After eight weeks different diets feeding, the stool was collected and the concentration of Akkermansia was analyzed. The result showed that the concentration of Akkermansia in HMD rats was lower than that of RD rats (Figure S2A). Furthermore, the serum levels of IL-6 and TNF-α in HMD rats were higher than that of RD rats after CLP models (Figure S2B). There were also higher number of total immune cells in lung in HMD rats after CLP model (Figure S2C). Therefore, methyl diets can change the intestinal flora and aggravate the degree of inflammatory reaction in CLP model rats. Thus, our results clearly showed that the higher inflammation and mortality in HMD mice or rats were through the alteration in gut microbiota composition.
Discussion
Sepsis has high morbidity and mortality and is the common cause of death in ICU,37 but there is no FDA-approved treatment option available for treating sepsis.38 For decades, antibiotics and supportive care have been commonly used to treat sepsis, but these treatments still cannot change the immune status of sepsis.39,40 The immune system, especially innate immune cells, plays a central role in the earlier phase of sepsis. Many studies, including ours, have shown that augmented proinflammatory response causes higher mortality.41,42 However, impaired cytokine and chemokine production was also shown to be related to higher mortality,7 which suggested that both excessive and impaired innate immune responses were associated with septic death. Although many regulators have been found to play important functions in the regulation of innate immune response and there is a growing understanding of the immune system response to sepsis, until now, there is still no effective therapy for treating sepsis.
It has been well known that DNA methylation is an epigenetic modification of the DNA process that can regulate DNA expression without changing the DNA sequence. In this study, we used a diet that is rich in methyl donors and cofactors (such as folic acid, betaine, cyanocobalamin, choline, and etc) to observe the effects of a high methyl diet on sepsis. We found that the high methyl diet exacerbated mortality and inflammation response in the CLP-induced septic model (Figure 1). However, the diet that is rich in methyl donors did not significantly affect the inflammatory response in LPS- or E.coli-induced septic model (Figures 2–3). We further confirmed this by LPS-stimulated in vitro macrophages activation and found that there was no significant difference in the activation of LPS-stimulated macrophages from HMD- and RD-fed mice. Therefore, dietary supplementation with methyl donors did not alter the inflammatory response of immune cells to the infection. To further analyze whether the innate immune cell responses are different between HMD and RD groups, we then examined the responses of LPS-treated macrophages from HMD- and RD-fed mice. There was no significant differences in IL-6 and TNF-α levels both in mRNA and protein levels (Figure 4), which suggests that the comparable innate immune cell response might not be the main cause of the higher CLP-induced mortality in HMD mice.
Gut functionality and microbial diversity have been shown to play an important role in sepsis pathogenesis,9,12,13 in regulating immune function and gut barrier integrity in the entire sepsis process. Studies have shown the change of gut microbiota could increase sepsis susceptibility.9,39,43 During the sepsis process, gut microbiota enhances the susceptibility of sepsis-induced organ dysfunction.8,39 Therefore, the disrupted gut microbiota has been shown to be related to the poor prognosis of sepsis.2 Studies have also shown that gut microbiota might be the new effective therapy for sepsis.2,33,39,44–46 Diet plays an important role in regulating gut microbiota and sepsis.6 Several pieces of research have demonstrated that different diet experiences affected the inflammatory response and mortality in sepsis.6,47–49 Previously published studies have tested methyl donor dietary supplementation in several inflammatory models,50 but none of them has reported that rich methyl donors altered the composition of gut microbiota. Consequently, we further detected the microbiome composition with 16S rRNA sequencing, followed by α-diversity and β-diversity analysis, and revealed that both HMD and RD dramatically altered microbiota composition in fecal (Figure 5). The lower abundance of probiotic Akkermansia and Lachnospiraceae_NK4A136_group was found in HMD mice.
Studies have shown that Akkermansia is involved in beneficial metabolite51 and is associated with several diseases, such as obesity, diabetes, and alcoholic liver disease.51–54 Our result showed that the percent of Akkermansia in total gut microbiota is significantly decreased to 0.0008829% in the HMD group whereas 32.47% in the RD group. Akkermansia was also shown to be associated with intestinal immune responses.55 Morowitz and colleagues found that non-fermentable fiber supplementation increased the abundance of Akkermansia and protected CLP-induced septic survival.6 The Lachnospiraceae family has health-promoting functions.29 Lachnospiraceae_NK4A136_group has been shown to play important functions in ulcerative colitis, diabetes, immune response, and the nutrients metabolism.36,56–59 To investigate whether HMD induces higher CLP-induced septic mortality through the alteration of gut microbiota, we transplanted feces from HMD mice into HMD and RD mice. Results showed that the transfer of feces, which were obtained from HMD mice, aggravated disease in recipient mice (Figure 6). These results confirmed that altered gut microbiota caused excessive inflammation in HMD mice.
Although methyl-related nutrients are shown to be good for health, our study showed that a high methyl diet exacerbated septic mortality and microbiome composition in mice. By adopting a fecal microbiome transplantation experiment, we demonstrated that the high methyl diet enhanced sepsis mortality through altering the gut microbiota. We found the abundance of bacteria with health-promoting functions like Akkermansia and Lachnospiraceae in the gut were decreased, and how did the HMD reduce these bacteria needs to be further explored. Our study showed that diet-based gut microbiota may be a new therapeutic strategy for sepsis patients.
Conclusions
In conclusion, a high methyl diet is able to alter the composition of gut microbiota and, therefore, exacerbate the mortality, organ injuries, and circulating levels of inflammatory mediators of CLP-induced septic mice.
Abbreviations
CLP, cecal ligation and puncture; DNA, deoxyribonucleic acid; EAE, experimental autoimmune encephalomyelitis; FMT, fecal microbiome transplantation; H&E, hematoxylin and eosin; HMD, high methyl diet; IBD, inflammatory bowel disease; IgA/G, immunoglobulin A/G; IL-6, interleukin-6; LPS, lipopolysaccharide; MPO, myeloperoxidase; MS, methionine synthase; PCA, principal component analysis; PCoA, principal co-ordinates analysis; RD, regulator diet (RD); TNF-α, tumor necrosis factor α.
Data Sharing Statement
All data and materials supporting the conclusions were included in this paper. More details are available on request.
Ethics Approval and Consent to Participate
All procedures involving animals were approved and performed in accordance with the Animal Care and Use Committee of Shanghai University of Chinese Traditional Medicine.
Acknowledgments
We would like to expand our heartfelt thanks to Dr. Zhiling Gao for sharing her experience of short-chain fatty acid sequencing and helping us in data analysis.
Author Contributions
All authors made substantial contributions to conception and design, acquisition of data, or analysis and interpretation of data; took part in drafting the article or revising it critically for important intellectual content; agreed to submit to the current journal; gave final approval of the version to be published; and agree to be accountable for all aspects of the work.
Funding
This work was supported by the National Natural Science Foundation of China (82070633 and 81873582 to X Kong, 81774256 to X Zhu, 31870905 to H Wu, 82074336 to X Sun, 81904164 to C Zhen), and Program of Shanghai Academic/Technology Research Leader (20XD1403700 to X Kong), and Si Ming Foundation of Shuguang Hospital Affiliated to Shanghai University of Traditional Chinese Medicine (SGKJ-201914 to C Yu).
Disclosure
The authors declare that they have no competing interests.
References
1. Singer M, Deutschman CS, Seymour CW, et al. The Third International Consensus Definitions for Sepsis and Septic Shock (Sepsis-3). JAMA. 2016;315(8):801–810. doi:10.1001/jama.2016.0287
2. Haak BW, Wiersinga WJ. The role of the gut microbiota in sepsis. Lancet Gastroenterol Hepatol. 2017;2(2):135–143. doi:10.1016/S2468-1253(16)30119-4
3. Reinhart K, Daniels R, Kissoon N, Machado FR, Schachter RD, Finfer S. Recognizing Sepsis as a Global Health Priority - A WHO Resolution. N Engl J Med. 2017;377(5):414–417. doi:10.1056/NEJMp1707170
4. Simonetto DA, Piccolo Serafim L, Gallo de Moraes A, Gajic O, Kamath PS. Gallo de Moraes A, Gajic O, Kamath PS. Management of Sepsis in Patients With Cirrhosis: current Evidence and Practical Approach. Hepatology. 2019;70(1):418–428. doi:10.1002/hep.30412
5. Fay KT, Klingensmith NJ, Chen C-W, et al. The gut microbiome alters immunophenotype and survival from sepsis. FASEB J. 2019;33(10):11258–11269. doi:10.1096/fj.201802188R
6. Morowitz MJ, Di Caro V, Pang D, et al. Dietary Supplementation With Nonfermentable Fiber Alters the Gut Microbiota and Confers Protection in Murine Models of Sepsis. Crit Care Med. 2017;45(5):e516–e523. doi:10.1097/CCM.0000000000002291
7. Liu W, Wu H, Chen L, Wen Y, Kong X, Gao WQ. Park7 interacts with p47(phox) to direct NADPH oxidase-dependent ROS production and protect against sepsis. Cell Res. 2015;25(6):691–706. doi:10.1038/cr.2015.63
8. Gong S, Yan Z, Liu Z, et al. Intestinal Microbiota Mediates the Susceptibility to Polymicrobial Sepsis‐Induced Liver Injury by Granisetron Generation in Mice. Hepatology. 2019;69(4):1751–1767. doi:10.1002/hep.30361
9. Singer JR, Blosser EG, Zindl CL, et al. Preventing dysbiosis of the neonatal mouse intestinal microbiome protects against late-onset sepsis. Nat Med. 2019;25(11):1772–1782. doi:10.1038/s41591-019-0640-y
10. Zeng MY, Cisalpino D, Varadarajan S, et al. Gut Microbiota-Induced Immunoglobulin G Controls Systemic Infection by Symbiotic Bacteria and Pathogens. Immunity. 2016;44(3):647–658. doi:10.1016/j.immuni.2016.02.006
11. Macpherson AJ, Koller Y, McCoy KD. The bilateral responsiveness between intestinal microbes and IgA. Trends Immunol. 2015;36(8):460–470. doi:10.1016/j.it.2015.06.006
12. Gallo RL, Hooper LV. Epithelial antimicrobial defence of the skin and intestine. Nat Rev Immunol. 2012;12(7):503–516. doi:10.1038/nri3228
13. Singer BH, Dickson RP, Denstaedt SJ, et al. Bacterial Dissemination to the Brain in Sepsis. Am J Respir Crit Care Med. 2018;197(6):747–756. doi:10.1164/rccm.201708-1559OC
14. Armacki M, Trugenberger AK, Ellwanger AK, et al. Thirty-eight-negative kinase 1 mediates trauma-induced intestinal injury and multi-organ failure. J Clin Invest. 2018;128(11):5056–5072. doi:10.1172/JCI97912
15. Lankelma JM, van Vught LA, Belzer C, et al. Critically ill patients demonstrate large interpersonal variation in intestinal microbiota dysregulation: a pilot study. Intensive Care Med. 2016;43(1):59–68. doi:10.1007/s00134-016-4613-z
16. Chassaing B, Koren O, Goodrich JK, et al. Dietary emulsifiers impact the mouse gut microbiota promoting colitis and metabolic syndrome. Nature. 2015;519(7541):92–96. doi:10.1038/nature14232
17. Bana B, Cabreiro F. The Microbiome and Aging. Annu Rev Genet. 2019;53:239–261. doi:10.1146/annurev-genet-112618-043650
18. Sochocka M, Donskow-Lysoniewska K, Diniz BS, Kurpas D, Brzozowska E, Leszek J. The Gut Microbiome Alterations and Inflammation-Driven Pathogenesis of Alzheimer’s Disease-a Critical Review. Mol Neurobiol. 2019;56(3):1841–1851. doi:10.1007/s12035-018-1188-4
19. Kadayifci FZ, Zheng S, Pan Y-X. Molecular Mechanisms Underlying the Link between Diet and DNA Methylation. Int J Mol Sci. 2018;19(12):12. doi:10.3390/ijms19124055
20. Li W, Tang R, Ma F, Ouyang S, Liu Z, Wu J. Folic acid supplementation alters the DNA methylation profile and improves insulin resistance in high-fat-diet-fed mice. J Nutr Biochem. 2018;59:76–83. doi:10.1016/j.jnutbio.2018.05.010
21. Zhang Y, Kutateladze TG. Diet and the epigenome. Nat Commun. 2018;9(1):3375. doi:10.1038/s41467-018-05778-1
22. Dejager L, Pinheiro I, Dejonckheere E, Libert C. Cecal ligation and puncture: the gold standard model for polymicrobial sepsis? Trends Microbiol. 2011;19(4):198–208. doi:10.1016/j.tim.2011.01.001
23. Rittirsch D, Huber-Lang MS, Flierl MA, Ward PA. Immunodesign of experimental sepsis by cecal ligation and puncture. Nat Protoc. 2009;4(1):31–36. doi:10.1038/nprot.2008.214
24. Wenjun L. Park7 interacts with p47phox to direct NADPH oxidase-dependent ROS production and protect against sepsis. Cell Res. 2015
25. Dingemanse C, Belzer C, van Hijum SA, et al. Akkermansia muciniphila and Helicobacter typhlonius modulate intestinal tumor development in mice. Carcinogenesis. 2015;36(11):1388–1396. doi:10.1093/carcin/bgv120
26. Li M, Sun X, Zhao J, et al. CCL5 deficiency promotes liver repair by improving inflammation resolution and liver regeneration through M2 macrophage polarization. Cell Mol Immunol. 2019.
27. Ryu JK, Kim SJ, Rah SH, et al. Reconstruction of LPS Transfer Cascade Reveals Structural Determinants within LBP, CD14, and TLR4-MD2 for Efficient LPS Recognition and Transfer. Immunity. 2017;46(1):38–50. doi:10.1016/j.immuni.2016.11.007
28. Jeschke J, Bizet M, Desmedt C, et al. DNA methylation-based immune response signature improves patient diagnosis in multiple cancers. J Clin Invest. 2017;127(8):3090–3102. doi:10.1172/JCI91095
29. Sorbara MT, Littmann ER, Fontana E, et al. Functional and Genomic Variation between Human-Derived Isolates of Lachnospiraceae Reveals Inter- and Intra-Species Diversity. Cell Host Microbe. 2020;28(1):134–146. doi:10.1016/j.chom.2020.05.005
30. Agus A, Planchais J, Sokol H. Gut Microbiota Regulation of Tryptophan Metabolism in Health and Disease. Cell Host Microbe. 2018;23(6):716–724. doi:10.1016/j.chom.2018.05.003
31. Gao X, Cao Q, Cheng Y, et al. Chronic stress promotes colitis by disturbing the gut microbiota and triggering immune system response. Proc Natl Acad Sci U S A. 2018;115(13):E2960–E2969. doi:10.1073/pnas.1720696115
32. Lavelle A, Sokol H. Gut microbiota-derived metabolites as key actors in inflammatory bowel disease. Nat Rev Gastroenterol Hepatol. 2020;17(4):223–237. doi:10.1038/s41575-019-0258-z
33. Liu S, Rezende RM, Moreira TG, et al. Oral Administration of miR-30d from Feces of MS Patients Suppresses MS-like Symptoms in Mice by Expanding Akkermansia muciniphila. Cell Host Microbe. 2019;26(6):779–794. doi:10.1016/j.chom.2019.10.008
34. Man SM. Inflammasomes in the gastrointestinal tract: infection, cancer and gut microbiota homeostasis. Nat Rev Gastroenterol Hepatol. 2018;15(12):721–737.
35. Miyamoto J, Igarashi M, Watanabe K, et al. Gut microbiota confers host resistance to obesity by metabolizing dietary polyunsaturated fatty acids. Nat Commun. 2019;10(1):4007. doi:10.1038/s41467-019-11978-0
36. Cui HX, Zhang LS, Luo Y, Yuan K, Huang ZY, Guo Y. A Purified Anthraquinone-Glycoside Preparation From Rhubarb Ameliorates Type 2 Diabetes Mellitus by Modulating the Gut Microbiota and Reducing Inflammation. Front Microbiol. 2019;10:1423. doi:10.3389/fmicb.2019.01423
37. Rhee C, Dantes R, Epstein L, et al. Incidence and trends of sepsis in us hospitals using clinical vs claims data, 2009–2014. JAMA. 2017;318(13):1241–1249. doi:10.1001/jama.2017.13836
38. Delano MJ, Ward PA. Sepsis-induced immune dysfunction: can immune therapies reduce mortality? J Clin Invest. 2016;126(1):23–31. doi:10.1172/JCI82224
39. Adelman MW, Woodworth MH, Langelier C, et al. The gut microbiome’s role in the development, maintenance, and outcomes of sepsis. Critical Care. 2020;24:1. doi:10.1186/s13054-020-02989-1
40. Stanski NL, Wong HR. Prognostic and predictive enrichment in sepsis. Nat Rev Nephrol. 2019;16(1):20–31. doi:10.1038/s41581-019-0199-3
41. Kong X, Thimmulappa R, Kombairaju P, Biswal S. NADPH oxidase-dependent reactive oxygen species mediate amplified TLR4 signaling and sepsis-induced mortality in Nrf2-deficient mice. J Immunol. 2010;185(1):569–577. doi:10.4049/jimmunol.0902315
42. Hiruma T, Tsuyuzaki H, Uchida K, et al. IFN-beta Improves Sepsis-related Alveolar Macrophage Dysfunction and Postseptic Acute Respiratory Distress Syndrome-related Mortality. Am J Respir Cell Mol Biol. 2018;59(1):45–55. doi:10.1165/rcmb.2017-0261OC
43. Zhu H, Liu Y, Li S, et al. Altered gut microbiota after traumatic splenectomy is associated with endotoxemia. Emerg Microbes Infect. 2018;7(1):197. doi:10.1038/s41426-018-0202-2
44. Angurana SK, Bansal A, Singhi S, et al. Evaluation of Effect of Probiotics on Cytokine Levels in Critically Ill Children With Severe Sepsis: a Double-Blind, Placebo-Controlled Trial. Crit Care Med. 2018;46(10):1656–1664. doi:10.1097/CCM.0000000000003279
45. Rubio I, Osuchowski MF, Shankar-Hari M, et al. Current gaps in sepsis immunology: new opportunities for translational research. Lancet Infect Dis. 2019;19(12):e422–e436. doi:10.1016/S1473-3099(19)30567-5
46. Bassetti M, Bandera A, Gori A. Therapeutic Potential of the Gut Microbiota in the Management of Sepsis. Crit Care. 2020;24(1):105. doi:10.1186/s13054-020-2780-3
47. Fuke N, Nagata N, Suganuma H, Ota T. Regulation of Gut Microbiota and Metabolic Endotoxemia with Dietary Factors. Nutrients. 2019;11:10. doi:10.3390/nu11102277
48. Bailey MA, Holscher HD. Microbiome-Mediated Effects of the Mediterranean Diet on Inflammation. Adv Nutrition. 2018;9(3):193–206. doi:10.1093/advances/nmy013
49. Napier BA, Andres-Terre M, Massis LM, et al. Western diet regulates immune status and the response to LPS-driven sepsis independent of diet-associated microbiome. Proc Natl Acad Sci U S A. 2019;116(9):3688–3694. doi:10.1073/pnas.1814273116
50. Cordero P, Gomez-Uriz AM, Campion J, Milagro FI, Martinez JA. Dietary supplementation with methyl donors reduces fatty liver and modifies the fatty acid synthase DNA methylation profile in rats fed an obesogenic diet. Genes Nutr. 2012;8(1):105–113. doi:10.1007/s12263-012-0300-z
51. Hänninen A, Toivonen R, Pöysti S, et al. Akkermansia muciniphila induces gut microbiota remodelling and controls islet autoimmunity in NOD mice. Gut. 2018;67(8):1445–1453. doi:10.1136/gutjnl-2017-314508
52. Cani PD. Human gut microbiome: hopes, threats and promises. Gut. 2018;67(9):1716–1725. doi:10.1136/gutjnl-2018-316723
53. Depommier C, Everard A, Druart C, et al. Supplementation with Akkermansia muciniphila in overweight and obese human volunteers: a proof-of-concept exploratory study. Nat Med. 2019;25(7):1096–1103. doi:10.1038/s41591-019-0495-2
54. Grander C, Adolph TE, Wieser V, et al. Recovery of ethanol-induced Akkermansia muciniphila depletion ameliorates alcoholic liver disease. Gut. 2018;67(5):891–901. doi:10.1136/gutjnl-2016-313432
55. Ansaldo E, Slayden LC, Ching KL, et al. Akkermansia muciniphila induces intestinal adaptive immune responses during homeostasis. Science. 2019;364(6446):1179–1184. doi:10.1126/science.aaw7479
56. Wang C, Li W, Wang H, et al. Saccharomyces boulardii alleviates ulcerative colitis carcinogenesis in mice by reducing TNF-alpha and IL-6 levels and functions and by rebalancing intestinal microbiota. BMC Microbiol. 2019;19(1):246. doi:10.1186/s12866-019-1610-8
57. Li M, Li F, Lu Z, et al. Effects of TiO2 nanoparticles on intestinal microbial composition of silkworm, Bombyx mori. Sci Total Environ. 2020;704:135273. doi:10.1016/j.scitotenv.2019.135273
58. Manuel CR, Latuga MS, Ashby CR, Reznik SE. Immune tolerance attenuates gut dysbiosis, dysregulated uterine gene expression and high-fat diet potentiated preterm birth in mice. Am J Obstet Gynecol. 2019;220(6):596e591–596 e528. doi:10.1016/j.ajog.2019.02.028
59. Huang G, Zhang S, Zhou C, et al. Influence of Eimeria falciformis Infection on Gut Microbiota and Metabolic Pathways in Mice. Infect Immun. 2018;86:5. doi:10.1128/IAI.00073-18
 © 2021 The Author(s). This work is published and licensed by Dove Medical Press Limited. The full terms of this license are available at https://www.dovepress.com/terms.php and incorporate the Creative Commons Attribution - Non Commercial (unported, v3.0) License.
By accessing the work you hereby accept the Terms. Non-commercial uses of the work are permitted without any further permission from Dove Medical Press Limited, provided the work is properly attributed. For permission for commercial use of this work, please see paragraphs 4.2 and 5 of our Terms.
© 2021 The Author(s). This work is published and licensed by Dove Medical Press Limited. The full terms of this license are available at https://www.dovepress.com/terms.php and incorporate the Creative Commons Attribution - Non Commercial (unported, v3.0) License.
By accessing the work you hereby accept the Terms. Non-commercial uses of the work are permitted without any further permission from Dove Medical Press Limited, provided the work is properly attributed. For permission for commercial use of this work, please see paragraphs 4.2 and 5 of our Terms.