Back to Journals » Infection and Drug Resistance » Volume 14
Metagenomic Next-Generation Sequencing Assists in the Diagnosis of Gardnerella vaginalis in Males with Pleural Effusion and Lung Infection: A Case Report and Literature Review
Authors Wu S , Hu W, Xiao W , Li Y, Huang Y , Zhang X
Received 29 September 2021
Accepted for publication 25 November 2021
Published 7 December 2021 Volume 2021:14 Pages 5253—5259
DOI https://doi.org/10.2147/IDR.S337248
Checked for plagiarism Yes
Review by Single anonymous peer review
Peer reviewer comments 2
Editor who approved publication: Prof. Dr. Héctor Mora-Montes
Simin Wu,1 Weihua Hu,1 Wei Xiao,1 Yongxia Li,2 Yan Huang,1 Xu Zhang1
1Department of Respiratory Medicine, The First Affiliated Hospital of Yangtze University, Jingzhou, 434000, Hubei, People’s Republic of China; 2Department of Respiratory Medicine, The Second Affiliated Hospital of Kunming Medical University, Kunming, 650000, Yunnan, People’s Republic of China
Correspondence: Weihua Hu; Wei Xiao Tel +86-18163137571; +86-18972161798
Email [email protected]; [email protected]
Abstract: Gardnerella vaginalis is a pathogen responsible for bacterial vaginosis, which is commonly found in female vaginas and rarely causes infections outside the female genitalia. Here, we report the use of metagenomic next-generation sequencing (mNGS) to detect and confirm pulmonary infection and pleural effusion caused by G. vaginalis in a 47-year-old man. The patient’s symptoms and imaging improved after 2 weeks of oral ornidazole, and he was cured after 3 months. Overall, the findings of this case demonstrate that mNGS is a useful tool for diagnosis of unexplained lung infections and pleural effusions. Its effectiveness in rapid and accurate etiological diagnosis and monitoring of diseases can allow detection of the etiology of difficult cases that return negative results after traditional cultures.
Keywords: Gardnerella vaginalis, lung infection, metagenomic next-generation sequencing
Introduction
Gardnerella vaginalis, a pathogen widely known to cause female vaginitis, rarely infects male genitalia. Moreover, extra-genitalia infection caused by G. vaginalis is rarely reported. Next-generation gene sequencing is an effective tool for rapid diagnosis of infectious pathogens. Here, we report a case of a male patient with no immunodeficiency, presented with pleural effusion and lung infection, conditions that were finally diagnosed as Gardnerella vaginalis infection by mNGS.
Case Presentation
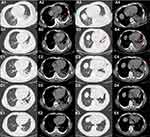
A 47-year-old man presented at the Emergency Clinic of The First Affiliated Hospital of Yangtze University, on October 5th 2020, with chest pain for 27 hours. The man, who worked at a lobster Catering Company, had a medical history of Heart Stent Implantation and hypertension, with a smoking index of 500. The patient felt a dull pain in the left chest at 9 am on the first day (October 4th), and this was accompanied by shortness of breath after activity. His chest pain was aggravated upon deep inhalation and turned over. Notably, although these symptoms were relieved by nitroglycerin, they reappeared about 1 to 2 hours later. On the second day, he went to the emergency department at 7 p.m. Electrocardiogram and troponin T examination revealed normal results. The patient was then referred from the chest pain center to the Department of Cardiology, where he was subjected to chest computed tomography (CT). Results revealed inflammation of lower lobes in both lungs and inferior lingular segment of the left lung, and left pleural effusion (Figure 1A1–A4). He was subsequently intravenously administered with ceftazidime (2 g), every 12 hours, for empirical anti-infection treatment. Physical examination showed that his consciousness was clear, the left lower lung was dulled in percussion, the left lung’s breath sounds were low, and no rale was heard in both lungs.
On October 7th 2020, he was transferred to the Department of Respiratory and Critical Care Medicine, where he was given an intravenous injection of levofloxacin (0.5 g) once a day. On October 9th, he was subjected to ultrasound-guided thoracentesis drainage and examination results of pleural effusion revealed that an adenosine deaminase (ADA) of 50.3 U/L, lactate dehydrogenase (LDH) of 2523 U/L, and protein level of 49.64 g/L. Neither bacteria nor tumor cells were found in pleural fluid cytology and pathology of the pleural effusion mass.
On October 10th, the antibiotics regimen was adjusted to ceftazidime and moxifloxacin, due to a rise in peak body temperature. On October 13th, this regimen was readjusted to meropenem and moxifloxacin due to lack of significant changes in the body temperature peak. Results from re-examination via chest CT on October 14th showed that double-under pneumonia was roughly the same, but the amount of pleural effusion was higher than before (Figure 1B1–B4). C reactive protein, erythrocyte sedimentation rate and blood routine examination were reexamined on October 15th and the changes are not obvious. On October 19th, we communicated with the patient, he voluntarily signed a written informed consent form, and he was subjected to bronchoscopy and his alveolar lavage collected. The alveolar lavage fluid was subjected to mNGS analysis targeting. mNGS was performed according to the standard protocol of the Illumina sequencing on the NextSeq550 platform. Bronchoscopy results revealed no abnormalities and his fever had disappeared. On October 21st, we received results of mNGS (see Table 1 for details): Summarily, sequence analysis revealed infection by G. vaginalis, and Corynebacterium urealyticum alongside other bacteria in the patient’s alveolar lavage fluid. Analysis of his sputum, blood and urine cultures during the hospitalization period all returned negative results.
 |
Table 1 Sequencing Results of mNGS from the Patient’s Alveolar Lavage Fluid Showing Detected Bacteria and Suspected Pathogens |
On October 23rd, the patient was discharged from hospital because of work reasons. Re-examination results from chest CT, performed on October 22nd prior to discharge, revealed that the pleural effusion in the left interlobular fissure tended to be wrapped, while the amount of effusion in the lower lobe of the left lung was slightly lower than before (Figure 1C1–C4). After discharge, he was instructed to regularly take ornidazole (0.5 g) tablets, orally twice a day (12th dose). On November 14th, 2020, the patient came to the clinic for review. Chest CT results showed that inflammation was basically absorbed in the right lower lobe and there was a slight reduction in the amount of interlobular fissure and pleural effusion on the left (Figure 1D1–D4). After review, he was again put on 2-week regimen of ornidazole tablets. Because the patient lived far away from the hospital, he requested a telephone follow-up. Later, he came to our outpatient clinic for physical examination on February 6th, 2021. Results indicated neither chest tightness nor discomfort, while CT-based re-examination of the chest showed that the inflammation in the lower lobe of the right lung was absorbed, and the inflammation on the left was basically absorbed (Figure 1E1–E4).
Discussion
This report describes a case of lung infection and pleural effusion caused by G. vaginalis and other vaginal flora, which was finally diagnosed by mNGS. Previous studies have shown that G. vaginalis, which mostly exists in female genitalia, is the main causative pathogen for bacterial vaginosis. The bacterium was discovered by Leopold,1 and was described as a “Haemophilus-like” species associated with cervicitis. G. vaginalis cells are gram-negative to gram-variable, small, pleomorphic rods that are nonmotile and do not possess flagella, endospores, or typical capsules.2 It can cause bacterial vaginitis, intrauterine infections, intra-amniotic infections, chorioamnionitis, postabortal pelvic inflammatory disease, and postpartum endometritis following cesarean section3 and it also exists in the vagina of healthy women.4 It is mainly diagnosed through secretion culture such as urine culture. Metronidazole is commonly used and gives initial cure rates of approximately 90% or better.2 Extravaginal infections by G. vaginalis such as bacteremia, joint infections, and spinal dural abscess, rarely occur,5–7 while lung infections and pleural effusions in female patients have also been documented.8 Similarly, studies have found that the vaginal Gardnerella biofilm is sexually transmitted, with some heterosexual couples found to share the same G. vaginalis strain,9 indicating that it can also be found in the male genitalia. However, reports about infection of other parts of men are relatively rare.
Literature from PubMed (https://www.ncbi.nlm.nih.gov/) inspired us to consider extravaginal infections caused by Gardnerella on three fronts. Firstly, this pathogen has been associated with bacteremia through bloodstream infections, which may cause infections in other parts of the body.6 Secondly, it has been associated with a specific subgroup of G. vaginalis. For example, Erica10 analyzed G. vaginalis subgroups in women who had sexual encounters with other women and found that different clades may have different pathogenicity levels, as well as different acquisition methods and infection durations, and it may spread among different groups of people or sexual networks. Thirdly, G. vaginalis exhibits numerous genetic diversity,11 and different genetic types/clades may have different levels of virulence. We consider that our patient’s infection may have been bacteremia, acquired through inhalation, or related to the bacterium’s biological characteristics, such as its subgroups, and specific genotypes, among others. Ultimately, we were unable to confirm the above views and route of infection, necessitating further explorations into the infection paths for this bacterium in men.
To date, 18 articles have reported G. vaginalis infections in men.12–29 Notably, destruction of the genitourinary tract is considered one of the predisposing factors for G. vaginalis urinary tract infections.22,23,25 In addition, the prostate is also considered a bacterial reservoir of G. vaginalis,12,16,19,20 while inflammation caused by prostate adenoma has been shown to promote passage of the body in the blood.16 At the same time, studies have reported two patients with pyelonephritis,13,20 while in patients with scrotal abscess, it is believed that the abscess is transmitted by cellulitis or infected hair follicles following infection with the relevant flora.28 Additional reports have also shown that sepsis is a common complication.18,20–22,27,28 The first reported case of G. vaginalis causing lung abscess, considering the aspiration bronchopneumonia caused by Gardnerella vaginalis with abscesses and signs of sepsis, although the infection route was not ultimately confirmed.15 In another case, G. vaginalis was detected in the patient’s alveolar lavage fluid, considering that the pathogen is likely to be able to attach to the lung epithelium, in a similar fashion to the way it attaches to the vaginal epithelium.26 Reports of lung infections caused by G. vaginalis are shown in the Table 2.
 |
Table 2 Reports of Lung Infections Caused by G. vaginalis |
Conventional methods of pathogenic diagnosis mostly involve culturing of infected samples, such as pleural fluid, urine or blood cultures. The disease diagnosis process used in the present case is outlined in Figure 2. We rarely take vaginal flora such as G. vaginalis into the consideration of possible pathogenic bacteria for the patient with pulmonary infection and pleural effusion. Since all our traditional culture results were negative, we employed mNGS to confirm etiological diagnosis.
 |
Figure 2 A flow chart describing the patient diagnosis process. |
To date, traditional culture methods remain the “gold standard” for detecting pathogens such as bacteria and fungi. However, these methods result in relatively low overall positive rates in blood cultures, at only 30% to 40%.30 mNGS has emerged as an efficient tool for analysis of genetic material obtained from microorganisms in patient samples. In fact, the technique is amenable to a wide range of microorganisms and is thus gradually being used for detection of pathogens across various clinical infectious diseases. Previous studies have shown that the turnaround time for second-generation gene sequencing from specimen receipt to completion of data analysis is approximately 6 hours to 7 days (48 hours on average), depending on the sequencing technology used, as well as the bioinformatics algorithms applied.31,32 Chen et al33 found that the diagnosis rate of bacteria and fungi using NGS was 95%, relative to only 60% obtained using culture method. Because NGS can detect dead bacteria, the use of effective empirical antibiotics may return positive NGS results of but negative ones for culture methods. On the other hand, mNGS guarantees efficient and accurate detection of etiology in the same specimen. Notably, compared to traditional cultivation, this technique confers higher accuracy for blood flow, respiratory infections, central nervous system, bone and joint infections, complex and atypical pathogen infections, thus improves precision diagnosis and treatment efficiency.32,34
To date, only a small number of respiratory pathogenic bacteria have been identified using traditional tests.35 We postulate that application of second-generation gene sequencing technology can improve the diagnosis rate of respiratory pathogens. For example, in the present case, we failed to diagnose G. vaginalis without applying mNGS. Notably, many pathogens in the environment and in the human body are difficult to cultivate, thus they may only be detected through mNGS.36 However, the clinical application of second-generation gene sequencing faces certain limitations. These include relatively high costs, high requirements for laboratories, lack of an internationally recognized interpretation guide to enable interpretation of results,37 and inability to distinguish between live from dead pathogens. In addition, the approach does not clearly prove the relationship between pathogens and the progress of the disease and has a relatively low detection efficiency for intracellular bacteria and fungi with cell walls.38
In conclusion, we reported the first case of use of mNGS to diagnose lung infection and pleural effusion caused by G. vaginalis. We envisage that this diagnostic technique will be further improved for better clinical application, especially for designing disease treatment strategies.
Consent
Written informed consent has been provided by the patient for the case details and images to be published. Details of the case can be published without institutional approval.
Disclosure
The authors report no conflicts of interest in this work.
References
1. Leopold S. Heretofore undescribed organism isolated from the genitourinary system. U S Armed Forces Med J. 1953;4(2):263–266.
2. Piot P. Gardnerella, streptobacillus, spirillum, and calymmatobacterium. In: Balows A, Hausler WJ
3. Catlin BW. Gardnerella vaginalis: characteristics, clinical considerations, and controversies. Clin Microbiol Rev. 1992;5(3):213–237. PMID: 1498765; PMCID: PMC358241. doi:10.1128/CMR.5.3.213
4. Zozaya-Hinchliffe M, Lillis R, Martin DH, Ferris MJ. Quantitative PCR assessments of bacterial species in women with and without bacterial vaginosis. J Clin Microbiol. 2010;48(5):1812–1819. doi:10.1128/JCM.00851-09
5. Stewart L, Sinha S, Madsen PJ, et al. Spinal epidural abscess caused by Gardnerella vaginalis and Prevotella amnii. Infect Dis Clin Pract. 2018;26(4):237. doi:10.1097/IPC.0000000000000565
6. Thomas M, Zeller V, Heym B, et al. Gardnerella vaginalis, from the vaginal microbiota to prosthetic joint infection. J Bone Joint Infect. 2019;4(4):189. doi:10.7150/jbji.32471
7. Hoarau G, Bernard S, Pavese P, et al. Gardnerella vaginalis as a rare cause of prosthetic joint infection. J Clin Microbiol. 2012;50(12):4154–4156. doi:10.1128/JCM.01969-12
8. Murray L, Halpin J, Casserly B, et al. A pyo-hydropneumothorax with sepsis, secondary to Gardnerella vaginalis infection in a post-partum female. Respir Med Case Rep. 2019;26:189–192. doi:10.1016/j.rmcr.2019.01.007
9. Swidsinski A, Doerffel Y, Loening-Baucke V, et al. Gardnerella biofilm involves females and males and is transmitted sexually. Gynecol Obstet Invest. 2010;70(4):256–263. doi:10.1159/000314015
10. Plummer EL, Vodstrcil LA, Murray GL, et al. Gardnerella vaginalis clade distribution is associated with behavioral practices and Nugent score in women who have sex with women. J Infect Dis. 2020;221(3):454–463. doi:10.1093/infdis/jiz474
11. Schellenberg JJ, Patterson MH, Hill JE. Gardnerella vaginalis diversity and ecology in relation to vaginal symptoms. Res Microbiol. 2017;168(9–10):837–844. doi:10.1016/j.resmic.2017.02.011
12. Patrick S, Gamett PA. Corynebacterium vaginale bacteraemia in a man. Lancet. 1978;1:987–998. doi:10.1016/S0140-6736(78)90273-8
13. Finkelhor RS, Wolinsky E, Kim CH, Tchou P, Frengley JD. Gardnerella vaginalis perinephric abscess in a transplant kidney. NEJM. 1981;304:846.
14. Wilson JA, Barratt AJ. An unusual case of Gardnerella vaginalis septicaemia. Br Med J. 1986;293(6542):309. doi:10.1136/bmj.293.6542.309
15. Legrand JC, Alewaeters A, Leenaerts L, et al. Gardnerella vaginalis bacteremia from pulmonary abscess in a male alcohol abuser. J Clin Microbiol. 1989;27(5):1132–1134. doi:10.1128/jcm.27.5.1132-1134.1989
16. Denoyel G-A, Drouet EB, Montclos HPD, et al. Gardnerella vaginalis bacteremia in a man with prostatic adenoma. J Infect Dis. 1990;161(2):367–368. doi:10.1093/infdis/161.2.367
17. Bastida-Vilá MT, López-Onrubia P, Rovira-Lledos J, Martinez-Martinez JA, Expósito-Aguilera M. Garderella vaginalis bacteremia in an adult male. Eur J Clin Microbiol Infect Dis. 1997;16:400–401. doi:10.1007/BF01726374
18. Calvert LD, Collins M, Bateman JRM. Multiple abscesses caused by Gardnerella vaginalis in an immunocompetent man. J Infect. 2005;51(2):E27–E29. doi:10.1016/j.jinf.2004.08.002
19. Lagacé-Wiens PRS, Ng B, Reimer A, et al. Gardnerella vaginalis bacteremia in a previously healthy man: case report and characterization of the isolate. J Clin Microbiol. 2008;46(2):804–806. doi:10.1128/JCM.01545-07
20. Yoon HJ, Chun J, Kim JH, Kang SS, Na DJ. Gardnerella vaginalis septicaemia with pyelonephritis, infective endocarditis and septic emboli in the kidney and brain of an adult male. Int J STD AIDS. 2010;21:653–657. doi:10.1258/ijsa.2010.009574
21. Alidjinou E, Bonnet I, Canis F, Dewulf G, Mazars E, Cattoen C. [Bactériémie à. Gardnerella vaginalis chez un homme]. Med Mal Infect. 2013;43:434–437. French. doi:10.1016/j.medmal.2013.07.007
22. Babics A, Roussellier P. Gardnerella vaginalis: an overlooked pathogen in male patients? Med Mal Infect. 2015;45(10):423–424. doi:10.1016/j.medmal.2015.09.007
23. Pritchard H. A case of pyelonephritis with bacteremia caused by Gardnerella vaginalis in a man. Infect Dis Clin Pract. 2018;26(6):e61–e63. doi:10.1097/IPC.0000000000000622
24. Bhatia P, Temple J, Kantor M. Not your garden-variety bacteremia: gardnerella in an immunocompromised man. Clin Infect Dis. 2018;66:1458–1459. doi:10.1093/cid/cix1054
25. Bittar J, Gazzetta J. Gardnerella vaginalis causing lung infection in young adult: a novel case. Respir Med Case Rep. 2019;28:100916. doi:10.1016/j.rmcr.2019.100916
26. Gómez ML, Way DA, PérezRamírez MD, Fernández JG. [Infecciones profundas por Gardnerella vaginalis en el varón. Revisión de la literatura y a propósito de un caso]. Rev Esp Quimioter. 2019;32(5):469–472. Spanish.
27. Alfraji N, Douedi S, Akoluk A, et al. Gardnerella vaginalis bacteremia in an elderly healthy male. IDCases. 2020;21:e00807. doi:10.1016/j.idcr.2020.e00807
28. Bekasiak A, Dammann F, Nader C. A rare cause of a scrotal abscess due to the symbiotic infection of Gardnerella vaginalis and Prevotella bivia in an adult male. Pathogens. 2020;9(2):93. doi:10.3390/pathogens9020093
29. Tamlihat YA, Augereau PF, Lardillon G, et al. Gardnerella vaginalis is a rare cause of ventilator-associated pneumonia: a case report and literature review. J Med Cases. 2021;12(4):134–137. doi:10.14740/jmc3645
30. Burillo A, Bouza E. Use of rapid diagnostic techniques in ICU patients with infections. BMC Infect Dis. 2014;14(1):593. doi:10.1186/s12879-014-0593-1
31. Pendleton KM, Erb-Downward JR, Bao Y, et al. Rapid pathogen identification in bacterial pneumonia using real-time metagenomics. Am J Respir Crit Care Med. 2017;196(12):1610–1612. doi:10.1164/rccm.201703-0537LE
32. Brown JR, Bharucha T, Breuer J. Encephalitis diagnosis using metagenomics: application of next generation sequencing for undiagnosed cases. J Infect. 2018;76(3):225–240. doi:10.1016/j.jinf.2017.12.014
33. Chen P, Sun W, He Y. Comparison of the next-generation sequencing (NGS) technology with culture methods in the diagnosis of bacterial and fungal infections. J Thorac Dis. 2020;12(9):4924.
34. Li N, Cai Q, Miao Q, et al. High‐throughput metagenomics for identification of pathogens in the clinical settings. Small Methods. 2020:2000792. doi:10.1002/smtd.202000792
35. Thorburn F. The Use of Next Generation Sequencing in the Diagnosis and Typing of Viral Infections. University of Glasgow; 2016.
36. Goldberg B, Sichtig H, Geyer C, et al. Making the leap from research laboratory to clinic: challenges and opportunities for next-generation sequencing in infectious disease diagnostics. MBio. 2015;6(6). doi:10.1128/mBio.01888-15
37. Dulanto CA, Dekker JP. From the pipeline to the bedside: advances and challenges in clinical metagenomics. J Infect Dis. 2020;221(Supplement_3):S331–S340. doi:10.1093/infdis/jiz151
38. Zhou X, Wu H, Ruan Q, et al. Clinical evaluation of diagnosis efficacy of active Mycobacterium tuberculosis complex infection via metagenomic next-generation sequencing of direct clinical samples. Front Cell Infect Microbiol. 2019;18(9):351. doi:10.3389/fcimb.2019.00351
 © 2021 The Author(s). This work is published and licensed by Dove Medical Press Limited. The full terms of this license are available at https://www.dovepress.com/terms.php and incorporate the Creative Commons Attribution - Non Commercial (unported, v3.0) License.
By accessing the work you hereby accept the Terms. Non-commercial uses of the work are permitted without any further permission from Dove Medical Press Limited, provided the work is properly attributed. For permission for commercial use of this work, please see paragraphs 4.2 and 5 of our Terms.
© 2021 The Author(s). This work is published and licensed by Dove Medical Press Limited. The full terms of this license are available at https://www.dovepress.com/terms.php and incorporate the Creative Commons Attribution - Non Commercial (unported, v3.0) License.
By accessing the work you hereby accept the Terms. Non-commercial uses of the work are permitted without any further permission from Dove Medical Press Limited, provided the work is properly attributed. For permission for commercial use of this work, please see paragraphs 4.2 and 5 of our Terms.