Back to Journals » OncoTargets and Therapy » Volume 10
Meta-analysis of the impact of de novo and acquired EGFR T790M mutations on the prognosis of patients with non-small cell lung cancer receiving EGFR-TKIs
Authors Liu Y, Sun L, Xiong Z, Sun X, Zhang S , Ma J, Han C
Received 24 January 2017
Accepted for publication 25 March 2017
Published 24 April 2017 Volume 2017:10 Pages 2267—2279
DOI https://doi.org/10.2147/OTT.S133082
Checked for plagiarism Yes
Review by Single anonymous peer review
Peer reviewer comments 3
Editor who approved publication: Dr Carlos E Vigil
Yang Liu, Li Sun, Zhi-Cheng Xiong, Xin Sun, Shu-Ling Zhang, Jie-Tao Ma, Cheng-Bo Han
Department of Oncology, Shengjing Hospital of China Medical University, Shenyang, People’s Republic of China
Purpose: The purpose of this meta-analysis was to explore the influences of pretreatment de novo and posttreatment-acquired epidermal growth factor receptor (EGFR) T790M mutations in patients with advanced non-small cell lung cancer (NSCLC) who had received tyrosine kinase inhibitors (TKIs).
Methods: We searched PubMed, Embase, and the China National Knowledge Infrastructure database for eligible literature. Data were extracted to assess the hazard ratios (HRs) for progression-free survival (PFS), overall survival (OS), and post-progression survival (PPS) and the relative ratios (RRs) for objective response rate (ORR).
Results: This meta-analysis included 22 studies comprising 1,462 patients with NSCLC who harbored activating EGFR mutations and were treated with EGFR-TKIs. Compared to pretreatment T790M mutation-negative NSCLC, pretreatment T790M mutation-positive NSCLC was associated with decreased PFS (HR 2.23, P<0.001) and OS (HR 1.55, P=0.003). A trend toward significance of worsening ORR (RR 0.86, P=0.051) was evident. The acquired T790M mutation was correlated with improved PFS (HR 0.75, P=0.006) and PPS (HR 0.57, P<0.001), compared to patients without the T790M mutation who progressed after EGFR-TKI treatment. There were no significant differences in OS or ORR between patients with acquired T790M mutation-positive and T790M mutation-negative NSCLC. However, in the tumor tissue rebiopsy subgroup, patients with acquired T790M mutation had improved OS (HR 0.60, P<0.001) compared to T790M mutation-negative patients. In the plasma ctDNA subgroup, acquired T790M mutation decreased the OS (HR 1.87, P<0.001).
Conclusion: Pretreatment T790M mutation was associated with worse PFS and OS in patients with advanced NSCLC treated with EGFR-TKIs, while acquired T790M mutation was associated with longer PFS and PPS than T790M mutation-negative NSCLC. The effects on OS were different between acquired T790M mutation detected from rebiopsy of tumor tissue and that detected from plasma ctDNA.
Keywords: epidermal growth factor receptor, T790M, non-small cell lung cancer, pretreatment, mutation
Background
Non-small cell lung cancer (NSCLC) accounts for more than 85% of lung cancers; half of the cases of NSCLC are classified as adenocarcinoma. Approximately 30%–50% of Asian and 10%–17% of Caucasian patients with lung adenocarcinoma harbor activating epidermal growth factor receptor (EGFR) mutations.1,2 EGFR-tyrosine kinase inhibitors (TKIs) such as gefitinib and erlotinib are the preferred treatment for patients with activating EGFR mutations (a deletion in exon 19 [19del] and a point mutation in exon 21 leading to substitution of leucine for arginine at position 858 [L858R]). Treatment with EGFR-TKIs achieves a significantly improved objective response rate (ORR) of 60%–80% and a progression-free survival (PFS) of 9–13 months, which are significantly improved outcomes compared to those achieved with chemotherapy.3,4
Nevertheless, most patients who initially respond to EGFR-TKIs will eventually acquire resistance. Approximately 50%–60% of the cases of resistance are mediated by a secondary T790M mutation (ie, a threonine-to-methionine substitution at amino acid position 790 in exon 20 [T790M]). The T790M mutation can induce steric hindrance to EGFR-TKIs and increase the affinity of the receptor to adenosine triphosphate, relative to its affinity to EGFR-TKIs, which abolishes the effect of EGFR-TKIs.5 AZD9291 (osimertinib [Tagrisso]) is the only third-generation EGFR-TKI approved by the US Food and Drug Administration for the treatment of acquired T790M mutation-positive advanced NSCLC after secondary resistance to first- or second-generation EGFR-TKIs. Significant improvements in PFS and ORR were observed in a phase I/II study of this drug.6–8 Some retrospective studies have observed that patients who experienced disease progression with or without acquired T790M mutation after EGFR-TKI therapy might have different prognoses.9–22 However, low rebiopsy rates and low sensitivities of detection methods after acquired resistance to EGFR-TKIs are challenging for clinical practice. Therefore, the development of noninvasive rebiopsy samples, such as plasma circulating tumor DNA (ctDNA), and high-sensitivity detection methods, such as digital polymerase chain reaction and next generation sequencing, is essential for monitoring dynamic changes in genes and selecting appropriate treatment strategies. Recently, the detection of EGFR mutations using plasma ctDNA and polymerase chain reaction-based or next generation sequencing methods has been confirmed as a feasible alternative strategy, if tumor tissue is not available. A moderate concordant rate of 65% in E20 T790M mutations between tumor and plasma ctDNA has been reported; this contrasts the high concordant rate of 90% in E19del and E21 L858R mutations.23,24
Recently, researchers have also explored the relationship between prognosis and pretreatment T790M mutation.25–32 Increasing evidence has indicated that T790M may exist at a low frequency within the tumor cells before EGFR-TKI treatment and may become the dominant clone only after drug selection pressure of EGFR-TKI treatment.25 Although reliable and widely accepted methods for detecting EGFR T790M mutation status have not yet been established, some researchers have attempted to detect T790M mutation before EGFR-TKI treatment using different assays with sensitivities ranging from 0.001% to 0.4%.25–32 This meta-analysis explored the influences of acquired T790M mutation following EGFR-TKI treatment and de novo T790M mutation prior to EGFR-TKI treatment on survival and prognosis in patients with advanced NSCLC who had activating EGFR mutations.
Methods
Literature search
PubMed, Embase, China National Knowledge Infrastructure database, and abstracts from major scientific meetings were searched for relevant articles published up to July 5, 2016. The following search terms were used: 1) lung cancer OR non-small cell lung cancer OR NSCLC; 2) T790M; and 3) progression-free survival (PFS) OR overall survival (OS) OR progression. The computer searches were supplemented with a manual search of the references listed in all retrieved review articles, primary studies, and meeting abstracts.
Study selection
Eligible studies for the pretreatment T790M group met several criteria. First, patients were confirmed to have advanced or recurrent NSCLC with activating EGFR mutations (19del or L858R mutation), and the status of the T790M mutation was detected before treatment with single-agent EGFR-TKI, that is, erlotinib or gefitinib (there was no limitation to the detection method). In the studies, EGFR-TKIs must have been used for the first time. Also, the study must have contained PFS or OS outcome data based on T790M mutation status; the corresponding hazards ratios (HRs) and 95% confidence intervals (CIs) could be directly obtained or calculated. Finally, PFS was defined as the time from the start of EGFR-TKI treatment to the first disease progression or death from any reason without progression; OS was defined as the time from the start of EGFR-TKI treatment or first diagnosis to the date of death by any cause or the date patients were last known to be alive. In all of the studies, the prevalence of T790M mutation was higher than 10%.
Eligible studies for the posttreatment-acquired T790M group met several criteria. First, patients were confirmed to have advanced or recurrent NSCLC before treatment with single-agent EGFR-TKI (erlotinib or gefitinib), and acquired resistance to EGFR-TKI was established according to the Jackman criteria33 (ie, patients who were EGFR wild-type or EGFR status unknown had an objective response [according to RECIST criteria] to EGFR-TKIs or had a period of durable stable disease [≥6 months] and eventually developed acquired resistance to EGFR-TKIs). Also, the status of the T790M mutation was detected after resistance to EGFR-TKIs following treatment with single-agent EGFR-TKI, that is, erlotinib or gefitinib (there was no limitation to the detection method). The study must have contained PFS, OS, or post-progression survival (PPS) outcome data; HRs and the corresponding 95% CIs for PFS, OS, and PPS based on T790M mutation status could be acquired or calculated. There was no upper limit for the number of lines of chemotherapy. Finally, PFS was defined as the time from the start of EGFR-TKI treatment to the first disease progression or death from any reason without progression; PPS was defined as time from the date of the first progression according to RECIST criteria version 1.1 to the second progression or death; and OS was defined as the time from the start of EGFR-TKI treatment to the date of death by any cause or the date patients were last known to be alive. In all of the studies, the prevalence of T790M mutation was higher than 10%.
Studies were excluded if they mentioned the use of third-generation EGFR-TKIs, repeated published studies, or included patients with small cell lung cancer. Studies were also excluded if patients simultaneously received other therapies or multiple targeted drug combinations. Finally, studies in which the data were insufficient and unable to meet the inclusion criteria were excluded.
Data extraction and quality assessment
The primary outcomes were PFS and OS, and the secondary outcomes were PPS and ORR. Two reviewers independently extracted author name, published date, total number of patients, method of EGFR detection, T790M mutation status, study outcomes (OS, PFS, PPS, and ORR) and the corresponding HR or relative ratio (RR), and patient characteristics. Discrepancies were discussed with a third investigator to reach an agreement. The Joanna Briggs Institute Prevalence Critical Appraisal Tool was used to assess the quality of the enrolled studies.34
Statistical analysis
All data were analyzed using the STATA 12.0 statistical software. The statistical heterogeneity of the enrolled studies was assessed using the inconsistency index (I2 statistic). If the I2 was >50% indicating significant heterogeneity, a random-effects model was used;35 otherwise, a fixed-effects model was used.36 HRs were extracted from the original studies or calculated from the reported number of events and the corresponding P-values of the log-rank statistics, as described by Tierney et al.37 The PFS, OS, and PPS were pooled and the results were analyzed according to HR and the corresponding 95% CI; ORR was pooled and the results were analyzed according to RR and the corresponding 95% CI. A P-value <0.05 was considered statistically significant for all analyses. Publication bias was examined with Begg and Mazumdar’s rank correlation test38 and Egger’s regression asymmetry test.39
Results
Search results
In total, the meta-analysis included 22 studies comprising 1,462 patients according to the inclusion and exclusion criteria. Figure 1 illustrates the selection process.
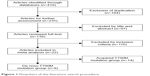  | Figure 1 Flowchart of the literature search procedure. |
Study characteristics
The pretreatment T790M mutation group included eight eligible studies involving 538 patients: 212 patients were T790M mutation positive and 326 patients were T790M mutation negative. The studies by Karachaliou et al28 and Rosell et al31 might share the same patients, despite the difference in survival-related data. Overall, seven studies including 447 patients reported PFStotal-related data, six studies including 420 patients reported PFSfirst-line-related data, four studies including 298 patients reported OStotal-related data, and five studies including 385 patients reported ORRtotal-related data. Table 1 lists the details of the studies included in the meta-analysis.
The posttreatment-acquired T790M mutation group included 14 eligible studies involving 924 patients: 445 patients were T790M mutation positive and 479 patients were T790M mutation negative. Three studies14,17,21 only provided initial data, so the P-values and corresponding HRs were calculated. Sorensen et al17 only reported patients who received second-line treatment with EGFR-TKIs. The study by Uramoto et al21 defined “TTP” as time-to-progression after gefitinib therapy, which was different from the definition in this meta-analysis, so TTP-related data were excluded. All the patients who had PPS-related data had rebiopsy tumor tissue specimens and almost received a rechallenge with TKIs (exclusive of third-generation TKIs) or standard chemotherapy regimens. Among the 14 studies included, nine studies including 405 patients reported PFS-related data, nine studies including 696 patients reported OS-related data, four studies including 282 patients reported PPS-related data, and five studies including 217 patients reported ORR-related data. Table 2 lists the details of the studies included in the analysis.
Quality assessment, heterogeneity analysis, and publication bias
The Joanna Briggs Institute Prevalence Critical Appraisal Tool was used to assess the quality of the enrolled studies (Tables 3 and 4). There was moderate heterogeneity in the pooled analysis of survival data (PFStotal, PFSfirst-line, OS, and ORR), and a random-effects model was used for final analysis (Table 5). Publication bias was assessed according to Egger’s and Begg’s regression methods and no significant publication bias was observed (P>0.05; Table 5).
  | Table 3 Quality assessment of studies included in the pretreatment T790M mutation group |
  | Table 4 Quality assessment of studies included in the posttreatment-acquired T790M mutation group |
Meta-analysis of pretreatment de novo T790M mutation
The rate of pretreatment T790M mutation-positive status ranged from 22.22% to 80% in the included studies. In the pretreatment T790M group, there was no significant heterogeneity between trials in the analysis of OStotal (I2=23.3%, P=0.271) or ORRtotal (I2=0.00%, P=0.499), so a fixed-effects model was used for analysis. A random-effects model was used when moderate heterogeneity existed between trials in the analysis of PFStotal (I2=60.6%, P=0.019) and PFSfirst-line (I2=56.5%, P=0.042). The heterogeneity did not decrease with the sensitivity analysis of the enrolled studies. Compared to T790M-negative patients, T790M-positive patients had significantly shorter PFStotal (HR 2.23, 95% CI 1.44–3.45, P<0.001; Figure 2A) and OStotal (HR 1.55, 95% CI 1.16–2.07, P=0.003; Figure 2B) in all treatment lines, as well as shorter PFSfirst-line (HR 1.91, 95% CI 1.23–2.97, P=0.004; Figure 3A) in first-line treatments. Additionally, pretreatment T790M-positive patients had a decreased ORRtotal compared to T790M-negative patients; there was a trend toward significance (RR 0.86, 95% CI 0.74–1.00, P=0.051; Figure 3B). Data showed that pretreatment T790M mutation may be predictive of worse survival in patients with NSCLC.
Meta-analysis of posttreatment-acquired T790M mutation
The rate of acquired T790M mutation-positive status after resistance to EGFR-TKI therapy ranged from 28% to 62.36% in the included studies. In the posttreatment-acquired T790M group, there was no significant heterogeneity between studies in the analysis of PFS (I2=30.2%, P=0.1.77) or PPS (I2=41.90%, P=0.16), so a fixed-effects model was used for analysis. The meta-analysis showed that T790M-positive patients had significantly longer PFS (HR 0.75, 95% CI 0.61–0.92, P=0.006; Figure 4A) and PPS (HR 0.57, 95% CI 0.44–0.73, P<0.001; Figure 4B) than T790M-negative patients. Heterogeneity was apparent in OS (I2=74.4%, P<0.001) and ORR (I2=66.9%, P=0.017) among the studies included, so a random-effects model was used. The heterogeneity of ORR did not change with the sensitivity analysis. The results showed that acquired T790M mutation-positive patients had similar OS (HR 0.86, 95% CI 0.55–1.36, P=0.526; Figure 5A) and ORR (RR 1.21, 95% CI 0.89–1.70, P=0.256; Figure 5B) compared to T790M-negative patients. PFS, OS, and ORR were further analyzed in two subgroups on the basis of type of rebiopsy specimen used for detection of EGFR mutation: tumor tissue or plasma ctDNA detection. In the tumor tissue rebiopsy subgroup, acquired T790M mutation significantly improved PFStissue (HR 0.76, 95% CI 0.59–0.98, P=0.037) and OStissue (HR 0.60, 95% CI 0.48–0.77, P<0.001) compared to T790M-mutation negative patients, but it did not increase ORRtissue (HR 0.93, 95% CI 0.85–1.49, P=0.759). In the plasma ctDNA subgroup, acquired T790M mutation significantly decreased OSctDNA (HR 1.87, 95% CI 1.49–2.36, P<0.001); no differences in PFSctDNA (HR 0.73, 95% CI 0.51–1.03, P=0.072) or ORRctDNA (HR 2.02, 95% CI 0.59–6.87, P=0.262) were observed between the two subgroups.
Discussion
The results of our meta-analysis indicate that pretreatment T790M mutation had a negative impact on PFS and OS in patients with NSCLC who harbored activating EGFR mutations and received EGFR-TKI treatment. In contrast, patients with acquired T790M mutation after resistance to EGFR-TKIs had significantly prolonged PFS and PPS, compared to patients without acquired T790M mutation. The subgroup analysis showed that OS benefit differed on the basis of the type of rebiopsy samples used for acquired T790M detection. In the tissue rebiopsy subgroup, OS was significantly improved, but, in the plasma ctDNA subgroup, OS was significantly inferior in patients with T790M mutation compared to those without T790M mutation.
Recently, highly sensitive genetic detection methods have been developed. Researchers are now largely able to identify pretreatment de novo T790M mutation existing at baseline before EGFR-TKI treatment. This achievement has attracted great interest related to drug sensitivity and survival prognosis. Pretreatment T790M mutation was reported to have no significant associations with the majority of the clinicopathologic characteristics such as age, stage, tumor size, or number of metastatic lymph nodes.40 However, patients with pretreatment T790M mutation tended to present with higher proportions of never-smoker status and brain metastasis.41 A previous meta-analysis by Ding et al42 indicated that pretreatment T790M mutation predicted inferior PFS in patients with NSCLC who harbored activating EGFR mutations and received EGFR-TKI treatment. Nevertheless, the relationship between pretreatment T790M mutation and OS has not been evaluated. This analysis revealed that pretreatment T790M mutation had a negative impact on OS. In a randomized phase III trial, patients with pretreatment T790M mutation-positive NSCLC had decreased PFS compared to patients with T790M mutation-negative NSCLC (9.7 vs 15.8 months, P=0.0185) when given erlotinib treatment.26 Among patients receiving chemotherapy, PFS was 6 months for T790M mutation-positive patients and 5.1 months for T790M mutation-negative patients (P<0.0001).11 Despite the prediction of poor prognosis for pretreatment T790M mutation, patients harboring activating EGFR mutations with or without the T790M mutation had longer PFS and better ORR compared to wild-type EGFR mutations when given EGFR-TKI therapy.25
It is not very clear why pretreatment T790M mutation predicts a negative effect on PFS and OS. A preclinical study showed that lung cancer cell lines with double-mutant T790M/L858R exhibited increased phosphorylated EGFR protein expression compared to cells with L858R mutation alone.43 When the T790M mutation was present in cells at a low percentage, the sensitivity to EGFR-TKI was similar to the sensitivity in cells harboring an activating EGFR mutation. When the T790M mutation in cells reached a certain percentage, the sensitivity to EGFR-TKIs obviously decreased.26,44 Clinical studies have shown that, with an increased abundance of pretreatment T790M mutation, patients had worse clinical outcomes in response to EGFR-TKI therapy.26,28,45 However, the European BELIEF study showed that patients with pretreatment T790M mutation benefitted more from erlotinib combined with bevacizumab than patients with T790M mutation-negative NSCLC (PFS 16.0 vs 10.5 months).46 Similarly, it is unclear whether patients harboring pretreatment T790M mutation will benefit more from third-generation EGFR-TKIs than patients without pretreatment T790M mutation. The ongoing FLAURA study might help to explore this important question.
Until now, research about the predictive role of acquired T790M mutation after EGFR-TKI therapy has been inconsistent. Rebiopsy of tumors after acquired resistance is vital to identify the mechanisms of resistance and choose subsequent therapy strategies. However, this is often not easily accomplished. In this meta-analysis, acquired T790M mutation after resistance to EGFR-TKI treatment predicted longer PFS, which was contrary to outcomes associated with the pretreatment T790M mutation. This could be explained by the fact that cells with acquired T790M mutation are characterized by indolent biologic behaviors;15,44 other complicated mechanisms of resistance to EGFR-TKIs might lead to patients being more refractory to subsequent treatment. In fact, the low abundance of pretreatment T790M mutation in tumor cells might gradually increase after EGFR-TKI therapy under selective pressure from drugs.25,27,47 One study involving 83 patients with activating EGFR mutations showed that patients with an increasing trend for T790M quantity from pretreatment to posttreatment of EGFR-TKIs had superior PFS and OS, compared to patients with a decreasing trend of T790M quantity.44,48 Thanks to recent advances in the era of third-generation EGFR-TKIs such as osimertinib, it is better understood that patients with acquired T790M mutation will benefit more from osimertinib treatment than patients without T790M mutation. Similar to resistance to first-generation EGFR-TKIs, acquired resistance to osimertinib is almost inevitable after a progression-free period of approximately 10 months. EGFR C797S, L718Q mutation, and amplification of HER-2, MET, and ERBB2 were found to be responsible for this resistance. Importantly, another drug, EAI045, has been developed to partially overcome the acquired resistance to AZD9291.49 In this analysis, all the included studies neither described the third generation of EGFR-TKIs nor mentioned that patients had ever received third-generation EGFR-TKI treatment. Therefore, our pooled results were not influenced by third-generation EGFR-TKI treatment. We believe acquired T790M mutation, relative to other resistance mechanisms after secondary resistance to EGFR-TKIs, suggests a better prognosis.
Several studies support these results. One study by Kuiper et al48 reported that acquired T790M mutation had a positive effect on PFS compared to T790M mutation-negative status after EGFR-TKI resistance (14.2 vs 11.1 months, P=0.034); no difference in OS was observed between the two arms (45.9 vs 29.8 months, P=0.213). Another study by Yu et al50 indicated that patients with acquired T790M mutation had improved PPS compared to T790M mutation-negative patients (1.9 vs 1.6 years, P=0.015). However, another study by Otsuka et al51 showed that acquired T790M mutation had a negative effect on PFS compared to T790M mutation-negative status after EGFR-TKI resistance (3.3 vs 4.1 months, P=0.048); no difference in OS was observed between the groups (15.1 vs 13.5 months, P=0.996). Interestingly, a randomized, controlled phase III trial (the IMPRESS study) indicated that patients who developed resistance to first-line gefitinib treatment failed to achieve benefits in PFS and OS from continuous gefitinib combined with chemotherapy compared to patients who received chemotherapy alone, regardless of T790M status.18 Other studies showed patients who progressed without the T790M mutation did benefit from the combination of continuous EGFR-TKIs and chemotherapy.9,13,15
This meta-analysis showed that acquired T790M mutation was not predictive of improved OS. Nevertheless, the subgroup analysis showed that OS was significantly superior in the tissue rebiopsy subgroup, but significantly inferior in the plasma ctDNA subgroup in patients with the T790M mutation compared to those without the T790M mutation. These findings suggest that high plasma levels of T790M mutation might be associated with an increased tumor burden, as well as tendencies for tumor progression and metastases.19,22 Thus, in order to individualize treatment, assessment of T790M status with both qualitative and quantitative analyses may be required. Combined detection of T790M in both tumor tissue and plasma ctDNA is a promising method for screening patients who might be appropriate candidates for osimertinib treatment.52,53
Limitations
There are some limitations to this meta-analysis. Significant heterogeneities were observed among the included studies. First, the enrolled studies used different EGFR detection methods with different sensitivities and specificities; these methods likely yielded false-negative and false-positive results.54,55 Second, the mutation tended to be heterogeneously distributed within the tumor tissue or plasma and some mutations (especially the T790M mutation) are only present in low proportions.56 In addition, the asymmetrical distribution of patient characteristics also influenced the results.
Conclusion
The clinical data included in this meta-analysis indicated that pretreatment T790M mutation was associated with worse PFS and OS in patients with advanced NSCLC who harbored activating EGFR mutations and received EGFR-TKI treatment, compared to patients without pretreatment T790M mutation. In contrast, acquired T790M mutation after resistance to EGFR-TKIs was associated with longer PFS and PPS, compared to T790M mutation-negative patients. Despite the fact that no significant difference was observed in OS in the total group, acquired T790M mutation detected from rebiopsy of tumor tissue had a positive effect on OS and mutation detected from plasma ctDNA had a negative effect on OS.
Disclosure
The authors report no conflicts of interest in this work.
References
Ettinger DS, Akerley W, Borghaei H, et al. Non-small cell lung cancer. J Natl Compr Canc Netw. 2012;10(10):1236–1271. | ||
Reguart N, Remon J. Common EGFR-mutated subgroups (Del19/L858R) in advanced non-small-cell lung cancer: chasing better outcomes with tyrosine kinase inhibitors. Future Oncol. 2015;11(8):1245–1257. | ||
Sun L, Ma JT, Zhang SL, Zou HW, Han CB. Efficacy and safety of chemotherapy or tyrosine kinase inhibitors combined with bevacizumab vs chemotherapy or tyrosine kinase inhibitors alone in the treatment of non-small cell lung cancer: a systematic review and meta-analysis. Med Oncol. 2015;32(2):473. | ||
Savas P, Hughes B, Solomon B. Targeted therapy in lung cancer: IPASS and beyond, keeping abreast of the explosion of targeted therapies for lung cancer. J Thorac Dis. 2013;5(Suppl 5):S579–S592. | ||
Kobayashi S, Boggon TJ, Dayaram T, et al. EGFR mutation and resistance of non-small-cell lung cancer to gefitinib. N Engl J Med. 2005;352(8):786–792. | ||
Jänne PA, Yang JC, Kim DW, et al. AZD9291 in EGFR inhibitor-resistant non-small-cell lung cancer. N Engl J Med. 2015;372(18):1689–1699. | ||
Yang J, Ramalingam SS, Jänne PA, Cantarini M, Mitsudomi T. LBA2_PR: osimertinib (AZD9291) in pre-treated pts with T790M-positive advanced NSCLC: updated Phase 1 (P1) and pooled Phase 2 (P2) results. J Thorac Oncol. 2016;11(4 Suppl):S152–S153. | ||
Wang S, Cang S, Liu D. Third-generation inhibitors targeting EGFR T790M mutation in advanced non-small cell lung cancer. J Hematol Oncol. 2016;9(1):34. | ||
Hata A, Katakami N, Yoshioka H, et al. Rebiopsy of non-small cell lung cancer patients with acquired resistance to epidermal growth factor receptor-tyrosine kinase inhibitor: Comparison between T790M mutation-positive and mutation-negative populations. Cancer. 2013;119(24):4325–4332. | ||
He C, Zheng L, Xu Y, Liu M, Li Y, Xu J. Highly sensitive and noninvasive detection of epidermal growth factor receptor T790M mutation in non-small cell lung cancer. Clin Chim Acta. 2013;425:119–124. | ||
Isobe K, Hata Y, Tochigi N, et al. Usefulness of nanofluidic digital PCR arrays to quantify T790M mutation in EGFR-mutant lung adenocarcinoma. Cancer Genomics Proteomics. 2015;12(1):31–37. | ||
Ji W, Choi CM, Rho JK, et al. Mechanisms of acquired resistance to EGFR-tyrosine kinase inhibitor in Korean patients with lung cancer. BMC Cancer. 2013;13:606. | ||
Li W, Ren S, Li J, et al. T790M mutation is associated with better efficacy of treatment beyond progression with EGFR-TKI in advanced NSCLC patients. Lung Cancer. 2014;84(3):295–300. | ||
Nakamura T, Sueoka-Aragane N, Iwanaga K, et al. A noninvasive system for monitoring resistance to epidermal growth factor receptor tyrosine kinase inhibitors with plasma DNA. J Thorac Oncol. 2011;6(10):1639–1648. | ||
Oxnard GR, Arcila ME, Sima CS, et al. Acquired resistance to EGFR tyrosine kinase inhibitors in EGFR-mutant lung cancer: distinct natural history of patients with tumors harboring the T790M mutation. Clin Cancer Res. 2011;17(6):1616–1622. | ||
Sakai K, Horiike A, Irwin DL, et al. Detection of epidermal growth factor receptor T790M mutation in plasma DNA from patients refractory to epidermal growth factor receptor tyrosine kinase inhibitor. Cancer Sci. 2013;104(9):1198–1204. | ||
Sorensen BS, Wu L, Wei W, et al. Monitoring of epidermal growth factor receptor tyrosine kinase inhibitor-sensitizing and resistance mutations in the plasma DNA of patients with advanced non-small cell lung cancer during treatment with erlotinib. Cancer. 2014;120(24):3896–3901. | ||
Soria JC, Wu YL, Nakagawa K, et al. Gefitinib/chemotherapy vs chemotherapy in EGFR mutation-positive NSCLC after progression on 1st line gefitinib (IMPRESS study): Final overall survival (OS) analysis. Ann Oncol. 2016;27(suppl_6):416–454. | ||
Sueoka-Aragane N, Katakami N, Satouchi M, et al. Monitoring EGFR T790M with plasma DNA from lung cancer patients in a prospective observational study. Cancer Sci. 2016;107(2):162–167. | ||
Sun JM, Ahn MJ, Choi YL, Ahn JS, Park K. Clinical implications of T790M mutation in patients with acquired resistance to EGFR tyrosine kinase inhibitors. Lung Cancer. 2013;82(2):294–298. | ||
Uramoto H, Yamada T, Yano S, Kondo N, Hasegawa S, Tanaka F. Prognostic value of acquired resistance-related molecules in Japanese patients with NSCLC treated with an EGFR-TKI. Anticancer Res. 2012;32(9):3785–3790. | ||
Zheng D, Ye X, Zhang MZ, et al. Plasma EGFR T790M ctDNA status is associated with clinical outcome in advanced NSCLC patients with acquired EGFR-TKI resistance. Sci Rep. 2016;6:20913. | ||
Oxnard GR, Thress KS, Alden RS, et al. 135O_PR: Plasma genotyping for predicting benefit from osimertinib in patients (pts) with advanced NSCLC. J Thorac Oncol. 2016;11(4 Suppl):S154. | ||
Zhou Q, Yang JJ, Chen ZH, et al. Serial cfDNA assessment of response and resistance to EGFR-TKI for patients with EGFR-L858R mutant lung cancer from a prospective clinical trial. J Hematol Oncol. 2016;9(1):86. | ||
Su KY, Chen HY, Li KC, et al. Pretreatment epidermal growth factor receptor (EGFR) T790M mutation predicts shorter EGFR tyrosine kinase inhibitor response duration in patients with non-small-cell lung cancer. J Clin Oncol. 2012;30(4):433–440. | ||
Costa C, Molina MA, Drozdowskyj A, et al. The impact of EGFR T790M mutations and BIM mRNA expression on outcome in patients with EGFR-mutant NSCLC treated with erlotinib or chemotherapy in the randomized phase III EURTAC trial. Clin Cancer Res. 2014;20(7):2001–2010. | ||
Fujita Y, Suda K, Kimura H, et al. Highly sensitive detection of EGFR T790M mutation using colony hybridization predicts favorable prognosis of patients with lung cancer harboring activating EGFR mutation.J Thorac Oncol. 2012;7(11):1640–1644. | ||
Karachaliou N, Costa C, Gimenez-Capita, et al. BRCA1, LMO4, and CtIP mRNA expression in erlotinib-treated non-small-cell lung cancer patients with EGFR mutations. J Thorac Oncol. 2013;8(3):295–300. | ||
Lee Y, Lee GK, Lee YS, et al. Clinical outcome according to the level of preexisting epidermal growth factor receptor T790M mutation in patients with lung cancer harboring sensitive epidermal growth factor receptor mutations. Cancer. 2014;120(14):2090–2098. | ||
Maheswaran S, Sequist LV, Nagrath S, et al. Detection of mutations in EGFR in circulating lung-cancer cells. N Engl J Med. 2008;359(4):366–377. | ||
Rosell R, Molina MA, Costa C, et al. Pretreatment EGFR T790M mutation and BRCA1 mRNA expression in erlotinib-treated advanced non-small-cell lung cancer patients with EGFR mutations. Clin Cancer Res. 2011;17(5):1160–1168. | ||
Zhao J, Feng HH, Zhao JY, et al. A sensitive and practical method to detect the T790M mutation in the epidermal growth factor receptor. Oncol Lett. 2016;11(4):2573–2579. | ||
Jackman D, Pao W, Riely GJ, et al. Clinical definition of acquired resistance to epidermal growth factor receptor tyrosine kinase inhibitors in non-small-cell lung cancer. J Clin Oncol. 2010;28(2):357–360. | ||
Munn Z, Moola S, Riitano D, Lisy K. The development of a critical appraisal tool for use in systematic reviews addressing questions of. Int J Health Policy Manage. 2014;3(3):123–128. | ||
DerSimonian R, Laird N. Meta-analysis in clinical trials. Control Clin Trials. 1986;7:177–188. | ||
Mantel N, Haenszel W. Statistical aspects of the analysis of data from retrospective studies of disease. J Natl Cancer Inst. 1959;22:719–748. | ||
Tierney JF, Stewart LA, Ghersi D, Burdett S, Sydes MR. Practical methods for incorporating summary time-to-event data into meta-analysis. Trials. 2007;8:16. | ||
Begg CB, Mazumdar M. Operating characteristics of a rank correlation test for publication bias. Biometrics. 1994;50(4):1088–1101. | ||
Egger M, Davey Smith G, Schneider M, Minder C. Bias in meta-analysis detected by a simple, graphical test. BMJ. 1997;315(7109):629–634. | ||
Oh JE, An CH, Yoo NJ, Lee SH. Detection of low-level EGFR T790M mutation in lung cancer tissues. APMIS. 2011;119(7):403–411. | ||
Lee Y, Lee GK, Hwang JA, Yun T, Kim HT, Lee JS. Clinical likelihood of sporadic primary EGFR T790M mutation in EGFR-mutant lung cancer. Clin Lung Cancer. 2015;16(1):46–50. | ||
Ding D, Yu Y, Li Z, Niu X, Lu S. The predictive role of pretreatment epidermal growth factor receptor T790M mutation on the progression-free survival of tyrosine-kinase inhibitor-treated non-small cell lung cancer patients: a meta-analysis. Onco Targets Ther. 2014;7:387–393. | ||
Mulloy R, Ferrand A, Kim Y, et al. Epidermal growth factor receptor mutants from human lung cancers exhibit enhanced catalytic activity and increased sensitivity to gefitinib. Cancer Res. 2007;67(5):2325–2330. | ||
Chmielecki J, Foo J, Oxnard GR, et al. Optimization of dosing for EGFR-mutant non-small cell lung cancer with evolutionary cancer modeling. Sci Transl Med. 2011;3(90):90ra59. | ||
Wang Z, Chen R, Wang S, et al. Quantification and dynamic monitoring of EGFR T790M in plasma cell-free DNA by digital PCR for prognosis of EGFR-TKI treatment in advanced NSCLC. PLoS One. 2014;9(11): e110780. | ||
Stahel AB, Dafni U, Gautschi O, et al. A phase II trial of erlotinib (E) and bevacizumab (B) in parents with advanced non-small-cell lung cancer (NSCLC) with activating epidermal growth factor receptor (EGFR) mutations with and without T790M mutation. The Spanish Lung Cancer Group (SLCG) and the European Thoracic Oncology Platform (ETOP) BELIEF trial. Eur J Cancer. 2015;51(Suppl 3):S711–S712. | ||
Nguyen KS, Kobayashi S, Costa DB. Acquired resistance to epidermal growth factor receptor tyrosine kinase inhibitors in non-small-cell lung cancers dependent on the epidermal growth factor receptor pathway. Clin Lung Cancer. 2009;10(4):281–289. | ||
Kuiper JL, Heideman DA, Thunnissen E, et al. Incidence of T790M mutation in (sequential) rebiopsies in EGFR-mutated NSCLC-patients. Lung Cancer. 2014;85(1):19–24. | ||
Wang S, Song Y, Yan F, Liu D. Mechanisms of resistance to third-generation EGFR tyrosine kinase inhibitors. Front Med. 2016;10(4):383–388. | ||
Yu H, Arcila ME, Rekhtman N, et al. Analysis of mechanisms of acquired resistance to EGFR TKI therapy in 155 patients with EGFR-mutant lung cancers. Clin Cancer Res. 2013;19(8):2240–2247. | ||
Otsuka K, Hata A, Takeshita J, et al. EGFR-TKI rechallenge with bevacizumab in EGFR-mutant non-small cell lung cancer. Cancer Chemother Pharmacol. 2015;76(4):835–841. | ||
Oxnard GR, Thress KS, Alden RS, et al. Association between plasma genotyping and outcomes of treatment with osimertinib (AZD9291) in advanced non-small-cell lung cancer. J Clin Oncol. 2016;34(28):3375–3382. | ||
Yang JC, Ahn MJ, Kim DW, et al. Osimertinib in pretreated T790M-positive advanced non-small-cell lung cancer: AURA Study Phase II Extension Component. J Clin Oncol. 2017:JCO2016703223. | ||
Ye X, Zhu ZZ, Zhong L, et al. High T790M detection rate in TKI-naive NSCLC with EGFR sensitive mutation: truth or artifact? J Thorac Oncol. 2013;8(9):1118–1120. | ||
Yu HA, Arcila ME, Hellmann MD, Kris MG, Ladanyi M, Riely GJ. Poor response to erlotinib in patients with tumors containing baseline EGFR T790M mutations found by routine clinical molecular testing. Ann Oncol. 2014;25(2):423–428. | ||
Nagai Y, Miyazawa H, Huqun, et al. Genetic heterogeneity of the epidermal growth factor receptor in non-small cell lung cancer cell lines revealed by a rapid and sensitive detection system, the peptide nucleic acid-locked nucleic acid PCR clamp. Cancer Res. 2005;65(16):7276–7282. |
 © 2017 The Author(s). This work is published and licensed by Dove Medical Press Limited. The full terms of this license are available at https://www.dovepress.com/terms.php and incorporate the Creative Commons Attribution - Non Commercial (unported, v3.0) License.
By accessing the work you hereby accept the Terms. Non-commercial uses of the work are permitted without any further permission from Dove Medical Press Limited, provided the work is properly attributed. For permission for commercial use of this work, please see paragraphs 4.2 and 5 of our Terms.
© 2017 The Author(s). This work is published and licensed by Dove Medical Press Limited. The full terms of this license are available at https://www.dovepress.com/terms.php and incorporate the Creative Commons Attribution - Non Commercial (unported, v3.0) License.
By accessing the work you hereby accept the Terms. Non-commercial uses of the work are permitted without any further permission from Dove Medical Press Limited, provided the work is properly attributed. For permission for commercial use of this work, please see paragraphs 4.2 and 5 of our Terms.