Back to Journals » International Journal of Nanomedicine » Volume 17
Mesenchymal Stem Cell Derived Exosomes as Nanodrug Carrier of Doxorubicin for Targeted Osteosarcoma Therapy via SDF1-CXCR4 Axis
Authors Wei H, Chen F, Chen J, Lin H, Wang S, Wang Y, Wu C, Lin J, Zhong G
Received 29 April 2022
Accepted for publication 27 July 2022
Published 4 August 2022 Volume 2022:17 Pages 3483—3495
DOI https://doi.org/10.2147/IJN.S372851
Checked for plagiarism Yes
Review by Single anonymous peer review
Peer reviewer comments 2
Editor who approved publication: Dr Farooq A. Shiekh
Hongxiang Wei,1,* Fei Chen,1,* Jinyuan Chen,2,* Huangfeng Lin,1 Shenglin Wang,1 Yunqing Wang,1 Chaoyang Wu,1 Jianhua Lin,1 Guangxian Zhong1
1Department of Orthopaedics, Fujian Institute of Orthopaedics, the First Affiliated Hospital of Fujian Medical University, Fuzhou, 350005, People’s Republic of China; 2The Centralab, the First Affiliated Hospital of Fujian Medical University, Fuzhou, 350005, People’s Republic of China
*These authors contributed equally to this work
Correspondence: Guangxian Zhong; Jianhua Lin, Tel/Fax +86 591 87981029, Email [email protected]; [email protected]
Purpose: The objective of this study was to investigate the antitumor activity, targeting capability, and mechanism of the developed nanodrug consisting of doxorubicin and exosome (Exo-Dox) derived from mesenchymal stem cells in vitro and in vivo.
Methods: The exosomes were isolated with Exosome Isolation Kit, and the Exo-Dox was prepared by mixing exosome with Dox-HCl, desalinizing with triethylamine and then dialyzing against PBS overnight. The exosome and Exo-Dox were examined by nanoparticle tracking analysis (NTA) and transmission electron microscopy (TEM). The antitumor activity, targeting capability, and mechanism of the developed Exo-Dox were evaluated by cell viability assay, histological and immunofluorescence analysis and in vivo imaging system.
Results: NTA results showed the size of the exosomes had increased from 141.6 nm to 178.1 nm after loading with doxorubicin. Compared with free Dox, the Exo-Dox exhibited higher cytotoxicity against osteosarcoma MG63 cells, HOS cells, and 143B cells than free Dox, the half-maximal inhibitory concentrations (IC50) of Dox, Exo-Dox were calculated to be 0.178 and 0.078 μg mL− 1 in MG63 cells, 0.294 and 0.109μg mL− 1 in HOS cells, 0.315 and 0.123 μg mL− 1 in 143B cells, respectively. The in vivo imaging showed that MSC derived Exo could serve as a highly efficient delivery vehicle for targeted drug delivery. The immunohistochemistry and histology analysis indicated that compared with the free Dox group, the Ki67-positive cells and cardiotoxicity in Exo-Dox group were significantly decreased.
Conclusion: Our results suggested that MSC-derived Exo could be excellent nanocarriers used to deliver chemotherapeutic drug Dox specifically and efficiently in osteosarcoma, resulting in enhanced toxicity against osteosarcoma and less toxicity in heart tissue. We further demonstrated the targeting capability of Exo was due to the chemotaxis of MSC-derived exosomes to osteosarcoma cells via SDF1-CXCR4 axis.
Keywords: targeted therapy, exosome, doxorubicin, osteosarcoma, nanocarrier
Introduction
Osteosarcoma is the most common malignant bone tumor that mainly occurs in children and adolescents.1–3 The standard treatments consist of preoperative neoadjuvant chemotherapy, surgical resection of osteosarcoma tissue, and postoperative chemotherapy with doxorubicin, cisplatin, and ifosfamide.4,5 Despite the fact that 5-year survival rate has improved from 20%-30% to 60%-70% with the introduction of neoadjuvant therapies,6 there are still some patients who fail chemotherapy due to the severe side effects which they cannot endure.7 Therefore, it is of great significance to develop targeted therapeutic agents with high selectivity and minimum side effects.
At present, the targeted drugs commonly used in clinic, such as Imatinib, Gefitinib, Trastuzumab, have no significant effect on osteosarcoma patients.8–10 Thus, a variety of nanocarriers for chemotherapeutic drugs have been considered a promising strategy in the targeted treatment of osteosarcoma.11–14 Generally, the nanocarriers can be classified into organic and inorganic carriers. The inorganic nanocarriers include metallic nanoparticles,15 mesoporous silica nanomaterials,16 carbon-based nanomaterials,17 and calcium phosphate carriers.18 And the organic nanocarriers mainly include liposomes,19 polymers,20 micelles,21 dendrimers,22 and exosomes.23 Specifically, exosome, as the natural membrane secreted by cells, was considered to be an ideal drug vehicle due to its excellent characteristics such as wide distribution in biological fluids, intrinsic homing capability, and its ability to penetrate the blood-brain barrier.24–26 Our group recently reported that bone marrow mesenchymal stem cell (BM-MSC) derived exomes could be used as a doxorubicin carrier for osteosarcoma treatment in vitro,27 however, the developed nanodrug’s (exosome-doxorubicin, Exo-Dox) therapeutic effect and targeting capability in vivo have not been elucidated.
In the present study, BM-MSCs, which have good performance such as tropism toward osteosarcoma tissue, low immunogenicity and side effects,28 were employed to produce exosomes for loading doxorubicin. Then, the developed nanodrug consisting of exosome-doxorubicin was characterized and administered to osteosarcoma loaded nude mouse model. The antitumor activity, targeting capability, and mechanism were further investigated in vitro and in vivo (Scheme 1).
Materials and Methods
Cell Lines and Culture
The human osteosarcoma cell line MG63, 143B, HOS and myocardial H9C2 cell line were purchased from the American Type Culture Collection (ATCC). The mouse bone marrow cells (BM-MSCs) with stable transfection of GFP were obtained from Cyagen Biosciences Inc (Guangzhou, China), the quality inspection report was done by the company. MSCs were maintained in MEM (Gibco Life Technologies, Grand Island, NY) containing 10% fetal bovine serum (FBS, ScienCell, San Diego, California, USA) and 1% penicillin–streptomycin (Gibco Life Technologies, USA). MG63, 143B, HOS and H9C2 cells were incubated in DMEM (Biological Industries, Israel) supplemented with 10% fetal bovine serum (FBS, Hyclone Thermo Scientific, USA) and 1% antibiotics. All cell lines were grown in an atmosphere containing 5% CO2 and maintained at 37°C in a humidified chamber (Thermo Fischer Scientific, Waltham, MA, USA).
Exosome Extraction, Drug-Loading and Identification
MSCs were washed and incubated for 48 h in mesenchymal stem cell medium-serum free (MSCM-sf medium, ScienCell, San Diego, California, USA) supplemented with 1% MSCGS-sf (ScienCell, cat. 7562) and 1% P/S Solution (ScienCell, cat. 0503) when they reached 70–80% confluence. The conditioned medium was collected using centrifugation (3000×g for 10 min) to discard cell debris. Then, the exosome suspension was mixed with Exosome Isolation Kit (Umibio, Shanghai, China) to isolate exosomes according to the manufacturer’s instructions. Next, the mixture was vortexed for 1 min and incubated for 2 h, followed by ultra-centrifugation for 60 min at 10,000 g after it was washed twice with a large volume of PBS and filtered using an Exosome Purification Filter (Umibio, Shanghai, China). The pelleted exosomes at the bottom of the tube were resuspended in PBS and centrifuged again at 12,000 g for 10 min to remove contaminating protein. All procedures were performed at 4°C and prepared exosomes were stored at −80°C until further experiments.
The preparation of Dox-loaded exosome (Exo-Dox) was based on the following steps. Briefly, the Doxorubicin hydrochloride (Dox HCl, Ourchem Shanghai, China) solution (1 mg mL−1) and purified exosome solution (1 mg mL−1) were mixed together for 30 min in a ratio of 7:93 and desalinized with triethylamine for 1 h at room temperature (RT). Then, the mixed solution was added to dialysis tube prepared by dialysis belt (cat. AT-C-026, Beijing, China) and centrifuge tube, and the excess Dox was removed by overnight dialysis in PBS solution at 4°C.
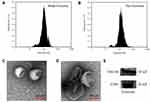
Isolated exosomes and prepared Exo-Dox were measured by nanoparticle tracking analysis (NTA) with Zeta View PMX 110 (Particle Metrix, Meerbusch, Germany) and corresponding software Zeta View 8.04.02 and visualized using a transmission electron microscope (TEM, HT7700, Hitachi, Japan). Western blot analysis was performed as follows: proteins of exosome were extracted using the Membrane and Cytosol Protein Extraction Kit (Beyotime, Shanghai, China), and their concentrations were quantified using the Bicinchoninic Acid Protein Assay Kit (Beyotime, Shanghai, China). After that, samples were electrophoresed on 10% sodium dodecyl sulfate-polyacrylamide gels and transferred to Immobilon membranes (Millipore, Bedford, MA, USA) and then incubated in QuickBlock™ Blocking Buffer (Beyotime, Shanghai, China) for 15 min. Then, using standard techniques, the membranes were probed with antibodies, including exosomal markers TSG101 (Affinity Biosciences, cat.DF8427) and CD81 (Affinity Biosciences, cat. DF2306). The membranes were then incubated with secondary antibodies (Affinity) at room temperature for 1 h. Labeling was visualized using the Affinity ECL Kit (Affinity, Biosciences, Cincinnati, USA) and the FluorChem R detection system (ProteinSimple, USA).
Cell Viability Assay
The proliferation of osteosarcoma cells and myocardial cells were measured using Cell Counting Kit-8 assay (New Cell & Molecular Biotech, Co., Ltd, China). MG63, HOS, 143B, and H9C2 cells were maintained at a density of 3000 cells/well in 96-well plates overnight and then treated with exosomes (0, 0.5, 1 2, 5, 10, 20, 50, 100 and 200 μg mL-1) and Dox formulations (Dox and Exo-Dox) with the equivalent Dox concentrations of 0, 0.01, 0.025, 0.05, 0.1, 0.25, 1, 5, 50, 10 μg mL−1for 48 h. Subsequently, the CCK-8 reagent was added to the medium, and the optical density (OD) at 450 nm was measured 2 h later.
Migration Assay
A migration assay was performed in 24-well transwell plates. The upper and lower culture compartments were separated by polycarbonate filters with 8-μm pore diameter (BD Biosciences, San Jose, CA). Equal numbers (50,000 cells) of MG63 cells (after treatment of free Exo, free Dox and Exo-Dox for 24 h) in serum free DMEM were added to the upper chamber. The lower chamber was filled with DMEM containing 10% FBS to induce cell migration, and plates were incubated for 24 h. Cells that traversed the membrane filter to the lower surface were fixed in 4% formalin for 15 min before staining with 0.1% crystal violet. Stained cells were counted directly using a microscope (Carl Zeiss Microscope Systems, Jena, Germany).
In vivo Anti-Tumor Efficacy
All experimental procedures were approved by the Committee of Animal Ethics of our hospital and conducted in accordance with the Guide for the Care and Use of Animals for research purposes. Nude mice (BALB/c, 4–6 weeks old) were purchased from SLAC, Shanghai, China. Xenograft mouse model was established by injecting MG63 cells (2 × 106 per mouse) into the left subcutaneous region of nude mice. Two weeks later, mice with similar tumor sizes were divided into four groups, and administered saline (control), free Exo, free Dox or Exo-Dox every 3 days x7, respectively. The dose was equivalent to 5 mg/kg Dox. Tumor sizes were measured every 3 days after intravenous injection of medicine. On day 21, the mice were sacrificed, and the hearts and tumor specimens were harvested. The tumor volume was calculated using the following formula: volume = length × width2/2).
Histological and Immunofluorescence Analysis
Pathological characterization of the collected rat specimens containing heart and tumor specimens was performed using a light microscope (Axioskop 40; Zeiss GmbH, Jena, Germany). Heart tissues were stained with hematoxylin-eosin (HE) to observe the cardiotoxicity of drugs, and tumor tissues were stained immunohistochemically with primary anti-Ki-67 Antibody (Affinity, Biosciences, Cincinnati, USA) guided by the instructions to investigate tumor cell proliferation. Two pathologists explored the immunohistochemical signals of specimens. The labeling index of Ki-67 expression was scored as 0 to 3 based on staining intensities: negative, 0; weakly positive, 1; moderately positive, 2; and strongly positive, 3. The mean percentage based on staining from 10 random high-power fields of positive tumor cells was also scored as 1 to 3 as follows: 1, <25%; 2, 25%–75%; and 3, >75%. Total scores were determined based on the intensity and percentage of positive staining in cancer cells. Overexpression was defined as a score >2, and low expression was defined as a score ≤2.
Osteosarcoma Tropism Mechanism of Exosomes
The total protein concentrations in cells and exosomes were determined using a BCA protein assay kit and equal amounts of protein were separated by SDS-PAGE gels and transferred to PVDF membranes. Antibodies against CXCR4, SDF1 and Tubulin were obtained from Affinity Biosciences (1:1000; Cincinnati, USA) and the negative marker calnexin was obtained from Thermo Scientific (Waltham, MA, USA). The immunoreactive bands were visualized using Affinity ECL Kit (Affinity Biosciences, Cincinnati, USA) and quantified using ImageJ software.
Construction of CXCR4 Knockout (KO) and Overexpression (OE) Cell Lines
The single guide RNAs (sgRNAs), plasmid vector, and empty vectors purchased from Ubigene (Guangzhou, China) were used to construct MG63 CXCR4 knockout and overexpression cell lines, denoted as MG63 CRCR4- or MG63 CXCR4+ cell line, respectively. For the disruption of the CXCR4 gene in osteosarcoma, the plasmids containing each target sgRNA sequence (sgRNA1 forward: 5ʹ-CACTTCAGATAACTACACCGAGG −3ʹ and sgRNA2 reverse: 5ʹ-GAAGCATGACGGACAAGTACAGG-3ʹ) and the corresponding negative control vectors were transfected into MG63 cells. Forty-eight hours later, stably transfected clones were screened using puromycin. Transfection efficiency was confirmed by Western blot analysis. The lentiviral packaging plasmid YOE-LV001-hCRCR4 (Ubigene, Guangzhou, China) was used to construct CXCR4 overexpression cell line according to the manufacturer’s instructions.
In vivo Imaging of Fluorescent Labeled Exosomes
MG63, CXCR4 knockout MG63 (MG63 CXCR4-) and CXCR4 overexpression MG63 (MG63 CXCR4+) cells were injected into male BALB/c nude mice to generate xenograft tumor models. Exosomes isolated from MSC cells were incubated with 1,1′-dioctadecyl-3,3,3′,3′-tetramethylindotricarbocyanine iodide (DiR) for 30 minutes. Saline (control), free DiR and Exo-DiR were injected intravenously into osteosarcoma bearing mice at a single DiR dose of 50 μg/kg. After anesthesia with isoflurane inhalation, the mice were scanned at different times (1, 3, 6, and 12 h) after injection using an in vivo imaging system (Bio-Real Sciences, Vienna, Austria) to detect the migration of fluorescent labeled exosomes in murine organs. The mice were euthanized at the final time point, and the major organs and tumors were extracted and immediately imaged again.
Statistical Analysis
All experiments were performed with at least three replicates per group. The statistical significance was determined using t-test and two-way analysis of variance with Tukey’s post hoc test. P values <0.05 were defined as statistically significant.
Results and Discussion
Characterization of the BM-MSC Derived Exosomes and Exo-Dox
The BM-MSCs were cultured in serum-free mesenchymal stem cell medium, supernatants were collected, and exosomes were extracted using Exosome Isolation Kit. As shown in Figure 1A and B, nanoparticle tracking analysis (NTA) showed the peak diameter of Exo and Exo-Dox was 141.6 and 178.1 nm, respectively, indicating increased diameter of Exo after loading with Dox. Transmission electron microscopy (TEM) revealed the typical lipid bilayer membrane encapsulated nanoparticles, and the diameter of Exo-Dox increased compared with free Dox, which was consistent with the results obtained by NTA (Figure 1C and D). Figure 1E demonstrated that the proteins of TSG101 and CD81 were positively expressed in BM-MSC derived exosomes.
In vitro Cytotoxicity and Antitumor Activity
The drug encapsulation efficiency, release profile, and cellular uptake of Exo-Dox in vitro were investigated in our previous work.27 In this study, the cytotoxicity of free Exo and the antitumor activity of Exo-Dox were further investigated after incubating with different cells for 48 h (Figure 2A and B). As shown in Figure 2A, the free Exo showed no significant cytotoxicity for osteosarcoma MG63 cells, HOS cells, 143B cells and myocardial H9C2 cells even at a high concentration of 200 ug mL−1, which demonstrated the excellent cytocompatibility of BM-MSC derived Exo.
As shown in Figure 2B, the Exo-Dox exhibited higher cytotoxicity against osteosarcoma MG63 cells, HOS cells, 143B cells than free Dox, the half-maximal inhibitory concentrations (IC50) of Dox, Exo-Dox were calculated to be 0.178 and 0.078 μg mL−1 in MG63 cells, 0.294 and 0.109 μg mL−1 in HOS cells, 0.315 and 0.123 μg mL−1 in 143B cells, respectively. However, in myocardial H9C2 cells, the Exo-Dox exhibited lower cytotoxicity than free Dox, the IC50 of Dox, Exo-Dox were calculated to be 0.149 and 0.461 μg mL−1, respectively. Moreover, considering that the osteosarcoma cells took about 24 hours to pass through a generation, we performed 24 h CCK-8 assay of MG63 and H9C2 cells. As shown in Figure S1A, no obvious cell death was observed after incubating with free exosome at the concentration of 200 μg mL−1 for 24 h, which indicated that the exosome had excellent cytocompatibility. In addition, as shown in Figure S1B, the IC50 (9.94μg mL-1 for Dox and 5.67μg mL−1 for Exo-Dox) for MG63 cells and (5.09μg mL−1 for Dox and 25.72μg mL−1 for Exo-Dox) for H9C2 cells were calculated automatically by using Graphpad prism 6.0, showing that the tumor cytotoxicity of Exo-Dox was stronger than free-Dox in MG63 cells, but in myocardial H9C2 cell line, the cytotoxicity of Exo-Dox was much weaker than that of free Dox, which was the main reason for using exosome as nanocarrier.
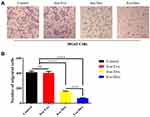
Transwell assay (Figure 3A and B) was carried out to investigate the effects of free Exo, free Dox, or Exo-Dox on migratory capacity of MG63 cells. As shown in Figure 3B, the treatment of free Dox suppressed migration to a degree (≈50%) compared with control group, and Exo-Dox treatment enhanced the suppression of MG63 migration compared with free Dox, whereas the treatment of free Exo did not cause an additional suppression. The results showed that Exo-Dox enhanced Dox-induced inhibition of MG63 cells’ migration. Collectively, from the results mentioned previously, we could see that the Exo-Dox could suppress proliferation and migration of osteosarcoma cells effectively and showed weakened cytotoxicity for myocardial cells, this may be attributed to the specific interaction of the membrane proteins in the surface of MSC-derived Exo and osteosarcoma cells.
Effect of Exo-Dox on Tumor Growth Inhibition in vivo
We further evaluated the in vivo therapeutic efficacy of Exo-Dox against MG63 cells in a xenograft nude mouse model. The tumor volumes and body weights of the mice were monitored at regular intervals. As shown in Figure 4A and B, the tumors of the control and free Exo groups progressed rapidly, indicating these two groups had no significant antitumor effect. However, the free Dox and Exo-Dox groups exhibited significant antitumor activity. Furthermore, at day 21 after drug administration, compared with the free Dox group, the Exo-Dox group had a 6-fold decrease in tumor volume. The mean tumor volume of Exo-Dox treated group was statistically significantly smaller than that of control group, free Exo group and free Dox group, respectively (P<0.01). In addition to changes in tumor volume, the tumor weight at the endpoint was also examined (Figure 4C and D). As shown in Figure 4D, the mean tumor weight of the free Dox group and Exo-Dox group were statistically significantly lower than that of control group and free Exo group, and the mean tumor weight of the Exo-Dox group was statistically significantly lower than that of free Dox group. Overall, these in vivo results showed synergistic effect of Exo and Dox against MG63 solid tumors, which was consistent with results obtained in vitro, indicating the Exo-Dox might preferentially interact with osteosarcoma cells.
Immunohistochemistry and Histology Analysis
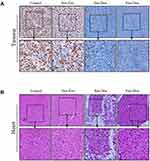
Immunohistochemical staining against Ki67 could be used to investigate tumor cell proliferation.29 As shown in Figure 5A, the Ki67-positive cells in control group and free Exo group were significantly more abundant than that in free Dox group and Exo-Dox group, suggesting that free Dox and Exo-Dox could effectively abrogate the tumor-proliferation rate. Moreover, compared with the free Dox group, the Ki67-positive cells in Exo-Dox group were significantly decreased, indicating the tumor inhibition was enhanced when using Exo-Dox. Furthermore, we found that the Ki67 level was not significantly altered between control group and free Exo group, showing that free Exo had no effect on tumor cell proliferation. These results were in accordance with the in vitro results of cell apoptosis assays, which could be explained by the fact that the membrane proteins in the surface of MSC-derived exosomes could specifically interact with the membrane proteins of MG63 cells.
Figure 5B showed the hematoxylin and eosin (H&E) staining of heart tissue after treating with different treatments. From Figure 5B, we can see that no obvious cardiac muscle tissue damage was observed in free Exo-treated groups compared with the control group, indicating that Exo had no effect on cardiac muscle tissue necrosis. In the free Dox group, the cardiomyocytes were significantly swollen, degenerated and necrotic, accompanied by vacuolar degeneration in the cytoplasm and rupture of myocardial fibers. However, compared with free Dox, Exo-Dox could significantly reduce the cardiotoxicity (Figure 5B), which could be ascribed to the chemotaxis of MSC-derived exosomes to osteosarcoma cells via SDF1-CXCR4 axis (Figure 6).
 |
Figure 6 The expression of CXCR4, SDF1 and tubulin proteins in 143B, MG63, HOS, H9C2, BM-MSCs cells and purified exosome measured by Western-blot analysis. |
The in vivo Imaging and Biodistribution of MSC-Derived Exo
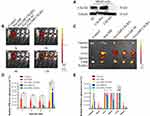
The tumor-targeting effect of MSC-derived Exo in vivo was evaluated by using a tumor xenograft mouse model (BALB/C nude mice). To further verify the targeted mechanism of of MSC-Exo, CXCR4 knockout MG63 (MG63 CXCR4-) and CXCR4 overexpression MG63 (MG63 CXCR4+) cells were also used to construct xenograft tumor models. Figure 7A indicated that the MG63 CXCR4- and MG63 CXCR4+ were successfully constructed.
Figure 7B shows the images obtained after live mice were scanned at 1 h, 3 h, 6 h, 12 h post-injection. As shown in Figure 7B and C, at 1 h after administration, the DiR signal was visualized mainly in the liver in free Dir group, Exo-DiR CXCR4+ group, Exo-DiR NC group and Exo-DiR group. At 3 h after administration, the fluorescence signal was observed in tumor regions of Exo-DiR CXCR4+ group and Exo-DiR NC group, then increased slightly at 6 h after administration. At 12 h after administration, there was strong fluorescence intensity in tumor regions of Exo-DiR CXCR4+ group and Exo-DiR NC group. However, the fluorescence intensity of tumor regions in control group, free DiR group and Exo-DiR CXCR4- group had no significant changes at 12 h after administration.
Overall, these results showed that MSC derived Exo could serve as a highly efficient delivery vehicle for targeted drug delivery. At 12 h after administration, the organs and tumors were harvested, and the fluorescence images were captured (Figure 7D and E). The images showed that in the tumor tissue, the fluorescence signals in Exo-DiR CXCR4+ group and Exo-DiR NC group were stronger than in other groups. In liver and lung tissue, there was strong fluorescence signal in all groups except control group. Whereas, in heart tissue, the fluorescence signals in Exo groups were weaker than in free DiR group. Collectively, these data suggested that MSC derived Exo performed well in targeted drug delivery to osteosarcoma tumors.
The results in Figure 7 indicated that BM-MSC exosomes could accumulate at the tumor site in vivo and could be used as a nanodrug carrier of doxorubicin to suppress proliferation and migration of osteosarcoma cells in vitro and in vivo (Figures 2–4). The mechanisms involved in the targeting effect of exosome for osteosarcoma were evaluated here. Osteosarcoma cells contain a lot of overexpressed surface proteins compared with normal cells.30 Owing to this characteristic, there might be specific enhanced receptor-mediated internalization of BM-MSC exosomes in osteosarcoma cells. Additionally, the mutual interaction of osteosarcoma cells and MSCs in metabolic reprograming has been studied.31,32 This evidence suggested that the cell membrane composition of exosomes may provide them with specific targeting properties. As shown in Figure 6, osteosarcoma cells highly expressed CXCR4 protein, while exosomes and its parent cell (mesenchymal stem cells) expressed SDF-1 protein. The interaction of SDF-1 and CXCR4 induced exosomes tending to the site of osteosarcoma. Thus, we speculated that mesenchymal stem cell derived exosomes exerted osteosarcoma targeting effect via SDF1-CXCR4 axis.
Conclusion
In conclusion, our data suggested that MSC-derived Exo could be used as excellent nanocarriers to deliver chemotherapeutic drug Dox specifically and efficiently to osteosarcoma, resulting in enhanced toxicity against osteosarcoma and less toxicity in heart tissue. Our results further showed that the mechanism of targeting capability of Exo could be ascribed to the chemotaxis of MSC-derived exosomes to osteosarcoma cells via SDF1-CXCR4 axis. This work may pave the road for new targeted approaches in osteosarcoma treatment.
Ethics Approval and Consent to Participate
All experimental procedures were approved by the Committee of Animal Ethics of the first Affiliated Hospital of Fujian Medical University and conducted in accordance with the Guide for the Care and Use of Animals for research purposes.
Acknowledgment
The authors gratefully acknowledge the financial support of the National Natural Science Foundation of China (81972021, 21904019), the Joint Funds for the innovation of science and Technology, Fujian province (2019Y9016), Fujian Province Science and Technology plan, China (2020Y4018), Natural Science Foundation of Fujian Province (2022J02031), the Middle-aged Backbone Project of the Fujian Provincial Health Commission (2021GGA034), the Excellent Talent Project of the First Affiliated Hospital of Fujian Medical University (YYXQN-ZGX2021), the special fund of youth top-notch innovative talents of Fujian Province (SQNBJ201601).
Disclosure
The authors report no conflicts of interest in this work.
References
1. Bishop MW, Janeway KA, Gorlick R. Future directions in the treatment of osteosarcoma. Curr Opin Pediatr. 2016;28(1):26–33. doi:10.1097/MOP.0000000000000298
2. Bao PP, Zhou YH, Lu WQ, et al. Incidence and mortality of sarcomas in Shanghai, China, during 2002-2014. Front Oncol. 2019;9:662. doi:10.3389/fonc.2019.00662
3. Corre I, Verrecchia F, Crenn V, Redini F, Trichet V. The osteosarcoma microenvironment: a complex but targetable ecosystem. Cells. 2020;9:976. doi:10.3390/cells9040976
4. Sayles LC, Breese MR, Koehne AL, et al. Genome-informed targeted therapy for osteosarcoma. Cancer Discov. 2019;9(1):46–63. doi:10.1158/2159-8290.CD-17-1152
5. Jafari F, Javdansirat S, Sanaie S, et al. Osteosarcoma: a comprehensive review of management and treatment strategies. Ann Diagn Pathol. 2020;49:151654. doi:10.1016/j.anndiagpath.2020.151654
6. Eaton BR, Schwarz R, Vatner R, et al. Osteosarcoma. Pediatr Blood Cancer. 2021;68(Suppl 2):e28352. doi:10.1002/pbc.28352
7. Zhang Y, Yang JQ, Zhao N, et al. Progress in the chemotherapeutic treatment of osteosarcoma (Review). Oncol Lett. 2018;16(5):6228–6237. doi:10.3892/ol.2018.9434
8. Zitvogel L, Rusakiewicz S, Routy B, Ayyoub M, Kroemer G. Immunological off-target effects of imatinib. Nat Rev Clin Oncol. 2016;13:431–446. doi:10.1038/nrclinonc.2016.41
9. Sanford M, Scott LJ. Gefitinib-A Review of its use in the treatment of locally advanced/metastatic non-small cell lung cancer. Drugs. 2009;69:2303–2328. doi:10.2165/10489100-000000000-00000
10. Modi S. Trastuzumab deruxtecan in previously treated HER2-positive metastatic breast cancer: plain language summary of the DESTINY-Breast01 study. Future Oncol. 2021;17(26):3415–3423. doi:10.2217/fon-2021-0427
11. Iram S, Zahera M, Khan S, et al. Gold nanoconjugates reinforce the potency of conjugated cisplatin and doxorubicin. Colloid Surface B. 2017;160:254–264. doi:10.1016/j.colsurfb.2017.09.017
12. Martinez-Carmona M, Lozano D, Colilla M, Vallet-Regi M. Lectin-conjugated pH-responsive mesoporous silica nanoparticles for targeted bone cancer treatment. Acta Biomater. 2018;65:393–404. doi:10.1016/j.actbio.2017.11.007
13. He K, Tang M. Safety of novel liposomal drugs for cancer treatment: advances and prospects. Chem Biol Interact. 2018;295:13–19. doi:10.1016/j.cbi.2017.09.006
14. Chi Y, Yin X, Sun K, et al. Redox-sensitive and hyaluronic acid functionalized liposomes for cytoplasmic drug delivery to osteosarcoma in animal models. J Control Release. 2017;261:113–125. doi:10.1016/j.jconrel.2017.06.027
15. Soleymaniha M, Shahbazi MA, Rafieerad AR, Maleki A, Amiri A. Promoting role of MXene nanosheets in biomedical sciences: therapeutic and biosensing innovations. Adv Healthc Mater. 2019;8:e1801137. doi:10.1002/adhm.201801137
16. Li T, Shi S, Goel S, et al. Recent advancements in mesoporous silica nanoparticles towards therapeutic applications for cancer. Acta Biomater. 2019;89:1–13. doi:10.1016/j.actbio.2019.02.031
17. Augustine S, Singh J, Srivastava M, Sharma M, Das A, Malhotra BD. Recent advances in carbon based nanosystems for cancer theranostics. Biomater Sci. 2017;5:901–952. doi:10.1039/c7bm00008a
18. Maia ALC, Ferreira CA, Barros ALB. Vincristine-loaded hydroxyapatite nanoparticles as a potential delivery system for bone cancer therapy. J Drug Target. 2018;26:592–603. doi:10.1080/1061186X.2017.1401078
19. Bulbake U, Doppalapudi S, Kommineni N, Khan W. Liposomal formulations in clinical use: an updated review. Pharmaceutics. 2017;9(2):12. doi:10.3390/pharmaceutics9020012
20. Haghiralsadat F, Amoabediny G, Naderinezhad S, Zandieh-Doulabi B, Forouzanfar T, Helder MN. Codelivery of doxorubicin and JIP1 siRNA with novel EphA2-targeted PEGylated cationic nanoliposomes to overcome osteosarcoma multidrug resistance. Int J Nanomed. 2018;13:3853–3866. doi:10.2147/IJN.S150017
21. Isoglu IA, Ozsoy Y, Isoglu SD. Advances in Micelle-based Drug Delivery: cross-linked Systems. Curr Top Med Chem. 2017;17:1469–1489. doi:10.2174/1568026616666161222110600
22. Wang H, Huang Q, Chang H, Xiao J, Cheng Y. Stimuli-responsive dendrimers in drug delivery. Biomater Sci. 2016;4:375–390. doi:10.1039/C5BM00532A
23. Palmerston Mendes L, Pan J, Torchilin VP. Dendrimers as Nanocarriers for Nucleic Acid and Drug Delivery in Cancer Therapy. Molecules. 2017;22(9):1401. doi:10.3390/molecules22091401
24. Simpson RJ, Jensen SS, Lim JW. Proteomic profiling of exosomes: current perspectives. Proteomics. 2008;8:4083–4099. doi:10.1002/pmic.200800109
25. Hood JL, San RS, Wickline SA. Exosomes released by melanoma cells prepare sentinel lymph nodes for tumor metastasis. Cancer Res. 2011;71:3792–3801. doi:10.1158/0008-5472.CAN-10-4455
26. Alvarez-Erviti L, Seow Y, Yin H, Betts C, Lakhal S, Wood MJA. Delivery of siRNA to the mouse brain by systemic injection of targeted exosomes. Nat Biotechnol. 2011;29:341–345. doi:10.1038/nbt.1807
27. Wei HX, Chen JY, Wang SL, et al. A nanodrug consisting of doxorubicin and exosome derived from mesenchymal stem cells for osteosarcoma treatment in vitro. Int J Nanomed. 2019;14:8603–8610. doi:10.2147/IJN.S218988
28. Kidd S, Spaeth E, Dembinski JL, et al. Direct evidence of mesenchymal stem cell tropism for tumor and wounding microenvironments using in vivo bioluminescent imaging. Stem Cells. 2009;27:2614–2623. doi:10.1002/stem.187
29. Jalava P, Kuopio T, Juntti-Patinen L, Kotkansalo T, Kronqvist P, Collan Y. Ki67 immunohistochemistry: a valuable marker in prognostication but with a risk of misclassification: proliferation subgroups formed based on Ki67 immunoreactivity and standardized mitotic index. Histopathology. 2006;48:674–682. doi:10.1111/j.1365-2559.2006.02402.x
30. Brune JC, Tormin A, Johansson MC, et al. Mesenchymal stromal cells from primary osteosarcoma are non-malignant and strikingly similar to their bone marrow counterparts. Int J Cancer. 2011;129:319–330. doi:10.1002/ijc.25697
31. Cortini M, Massa A, Avnet S, Bonuccelli G, Baldini N. Tumor-Activated Mesenchymal Stromal Cells Promote Osteosarcoma Stemness and Migratory Potential via IL-6 Secretion. PLoS One. 2016;11:e0166500–522. doi:10.1371/journal.pone.0166500
32. Mohammad IK, Reem KA, Hani C, Aamir A. Exosome-Mediated Response to Cancer Therapy: modulation of Epigenetic Machinery. Int J Mol Sci. 2022;23(11):6222. doi:10.3390/ijms23116222
 © 2022 The Author(s). This work is published and licensed by Dove Medical Press Limited. The full terms of this license are available at https://www.dovepress.com/terms.php and incorporate the Creative Commons Attribution - Non Commercial (unported, v3.0) License.
By accessing the work you hereby accept the Terms. Non-commercial uses of the work are permitted without any further permission from Dove Medical Press Limited, provided the work is properly attributed. For permission for commercial use of this work, please see paragraphs 4.2 and 5 of our Terms.
© 2022 The Author(s). This work is published and licensed by Dove Medical Press Limited. The full terms of this license are available at https://www.dovepress.com/terms.php and incorporate the Creative Commons Attribution - Non Commercial (unported, v3.0) License.
By accessing the work you hereby accept the Terms. Non-commercial uses of the work are permitted without any further permission from Dove Medical Press Limited, provided the work is properly attributed. For permission for commercial use of this work, please see paragraphs 4.2 and 5 of our Terms.