Back to Archived Journals » Open Access Bioinformatics » Volume 7 » Default
Mediation analysis in genome-wide association studies: current perspectives
Authors Lutz S, Hokanson J
Received 4 May 2015
Accepted for publication 28 July 2015
Published 11 November 2015 Volume 2015:7 Pages 1—5
DOI https://doi.org/10.2147/OAB.S63643
Checked for plagiarism Yes
Review by Single anonymous peer review
Peer reviewer comments 3
Editor who approved publication: Professor Charles Arntzen
Sharon M Lutz,1 John E Hokanson2
1Department of Biostatistics and Informatics, 2Department of Epidemiology, University of Colorado, Anschutz Medical Campus, Aurora, CO, USA
Abstract: In the course of genome-wide association studies (GWAS) over the last decade, genetic associations of the same locus with multiple phenotypes (eg, pleiotropy) have been observed. Pleiotropy may represent either common or distinct biological pathways. Mediation analysis provides insight into interpreting the pleiotropy identified through GWAS as either shared or distinct effects. In this paper, we will discuss mediation analysis for genetic effects, the challenges and limitations associated with these methods, and the importance of mediation analysis in the GWAS era.
Keywords: mediation analysis, genome-wide association studies, pleiotropy
Introduction
Genome-wide association studies (GWAS) are used to identify novel genetic associations. In the course of GWAS investigations over the last decade, genetic associations of the same locus with multiple phenotypes (eg, pleiotropy) have been observed. Pleiotropy may represent either common or distinct biological processes. Mediation analysis offers a strategy for identifying phenotypes that share a common genetic pathway and for quantifying the proportion of the total genetic effect on those phenotypes. Mediation analysis provides insight into interpreting the pleiotropy identified through GWAS as either shared or distinct effects.
Attempts have been made to identify common pathways for the observed pleiotropy found for the chromosome 15q25 region. Multiple GWAS have found significant signals in this region for clinical outcomes such as lung cancer,1 chronic obstructive pulmonary disease (COPD),2 and cigarette smoking.3,4 Within this chromosomal region, there is a cluster of nicotinic acetylcholine receptor genes CHRNA5/A3 that may have an indirect or direct effect on clinical disease (eg, COPD) through smoking behavior. However, there are other significant GWAS signals in this region for IREB2 and AGPHD1 that may directly influence lung biology.
Mediation analysis has been applied to this problem. Single-nucleotide polymorphisms (SNPs) in this chromosome 15q25 region for IREB2, AGPHD1, and CHRNA3 were identified from previous GWAS.5 Both direct and indirect effects of these SNPs were tested with cigarette smoking as a mediator of COPD. SNPs within the CHRNA3 and AGPHD1 regions show both direct effects and indirect effects on COPD mediated by smoking while IREB2 was not associated with smoking nor showed any indirect effect of smoking on COPD.5 The results of this mediation analysis indicate that the genetic susceptibility to COPD can be partitioned into pathways mediated by smoking behavior and pathways independent of current measures of smoking behavior.6
Mediation analysis has been used to examine pleiotropy in several other GWAS, including the following examples. A study has shown that an SNP on chromosome 14q21 and reduced bladder cancer risk are mediated by telomere length.7 Another mediation analysis revealed a suppression effect of the adiponectin levels on the association between CDH13 genotypes and metabolic syndrome.8 An additional study has shown that macronutrient intake is a mediator with FTO SNPs to increase body mass index.9 A recent study has shown that the AHSG gene is associated with bone mineral density through fetuin-A and body mass index.10
In addition to pleiotropy in GWAS, obvious biological mediators for a gene and clinical outcome are mRNA expression as measured by expression quantitative trait loci, DNA methylation, and the protein product of the gene.11,12 For instance, a recent study used mediation analysis to determine whether observed trans-expression quantitative trait locus associations are mediated by expression of transcripts in cis with the SNPs showing trans-association.13 Mediation analysis can provide crucial information regarding the potential mechanisms for observed pleiotropy from GWAS and in deciphering direct and indirect pathways responsible for the pleiotropic effects.
In this paper, we will discuss causal inference and mediation analysis for genetic effects, the challenges, assumptions, and limitations associated with these methods, and the importance of mediation analysis in GWAS.
Causal inference and mediation analysis for genetic effects
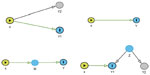
In GWAS, different complex phenotypes are often associated with the same genetic marker. Such associations can be indicative of pleiotropy (eg, common genetic causes),14 direct genetic effects via one of these phenotypes,15,16 indirect genetic effects via one or more of these phenotypes,17–19 or can be solely attributable to non-genetic/environmental links between the traits. These four conditions are illustrated in Figure 1. To identify the phenotypes with the inducing genetic association, statistical methodology is needed that is able to distinguish between the different causes of the genetic associations.19,20 Vansteelandt and Lange discuss statistical methodology and developments in the causal inference literature relevant to these analytical issues in genetic association studies, including counterfactuals and Directed Acyclic Graphs.19
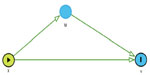
A subset of causal inference, mediation analysis quantifies the proportion of direct and indirect effects of the exposure on the outcome of interest via the mediator, an intermediate phenotype on the causal pathway between the exposure and the outcome.17,18 For example, mediation analysis can be used to determine what proportion of the total genetic effect that an SNP on chromosome 15 (CHRNA5/3) has on forced expiratory volume in 1 second (FEV1) (a measure of pulmonary function) is mediated through pack-years of smoking history (the putative intermediate phenotype). For mediation analysis, first consider the simple scenario for a continuous outcome Y (eg, FEV1) and mediator M (eg, pack-years of smoking history) as seen in Figure 2.
Three linear regression models can be used to jointly describe the effect of the SNP (X) on the outcome Y through M as seen in the following equations.21
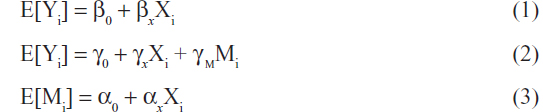
In Equation 1, βx represents the total effect of the exposure X on the outcome Y both through the mediator M and through other pathways. In Figure 2, βx measures both paths X → M → Y and X → Y. In equation 2, γx represents the direct effect of the exposure X on the outcome Y. In Figure 2, γx measures the path X → Y. In equation 2, γm represents the effect of the mediator M on the outcome Y adjusting for the exposure X. In Figure 2, γm measures the path M → Y. In Equation 3, αx represents the association between the exposure X and the mediator M and in Figure 2, αx measures the path X → M.
An indirect effect of X on Y through M occurs when in Equation (3) and in Equation (2). This is referred to as the intersection union test, causal steps approach,22 or the joint significance test.23,24 The indirect effect can be estimated as and tested using the Sobel test.25 These mediation models can be used to calculate the proportion of the direct and indirect effects of the exposure X on the outcome Y.26
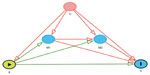
Mediation models have been extended from this simple scenario described above to accommodate interactions between the exposure and mediator,27,28 adjustment for confounders,16,17,29 assessment of direct and mediated effects,30–32 and multiple mediators.20 Figure 3 depicts one of these extended scenarios: a Directed Acyclic Graph with multiple mediators and confounders. These methods make several assumptions and have potential limitations when applied to GWAS to determine the effect of the SNP on the outcome through the mediator.
Assumptions and challenges
When using mediation analysis, one needs to be cognizant of the assumptions made for these methods to be valid. Most mediation methods assume there are no unmeasured confounding of the exposure–outcome relationship, no unmeasured confounders of the mediator–outcome relationship, and no unmeasured confounding of the exposure–mediator relationship.20 This may be an issue if there is ascertainment bias in case–control studies. For instance, an SNP may be associated with both the mediator and the outcome because the SNP affects the outcome and the outcome is correlated with the mediator. This may result in an indirect pathway that is merely the result of the sampling bias of the study.33,34 Several methods have been proposed that adjust for this ascertainment bias due to case control sampling.35–42
In addition, if there is measurement error of the exposure and/or the mediator, this can result in an incorrect interpretation.43 For instance, if there is non-differential misclassification of a binary mediator, then the natural direct effect may be overestimated and the natural indirect effect may be underestimated.44 Mediation analysis also assumes a causal direction of the effect that must come from a priori biological knowledge.19 For instance, mediation methods are assuming that the arrows in Figures 1–3 are correctly specified.
While there are methods that incorporate multiple mediators, one needs to consider the correlation between these mediators to avoid issues due to colinearity.20 While several mediators can be tested individually, a correction for multiple testing should be done if these mediators are measurements of the same phenotype. This correction for multiple comparisons can decrease power if an investigator uses several phenotypes that measure the same trait. For instance, consider the scenario where one wants to test whether the same SNP is associated with COPD through the mediator nicotine dependence and nicotine dependence is measured as pack-years of smoking history and current/former smoking status. There are two approaches to this problem. 1) One could use a method that accommodates multiple mediators assuming current smoking status is not heavily correlated with pack-years of smoking history in the study.20 2) One could test both whether the SNP is associated with COPD through pack-years of smoking history and whether the SNP is associated with COPD through current smoking status and then correct the alpha level for the two tests that were run. As a result, one needs to be careful when choosing the number of mediators to include in the analysis.
In general, mediation analysis is not run for every SNP in a GWAS since mediation analysis is unfounded if the SNP does not affect the outcome of interest. First, a GWAS is run for the outcome of interest (eg, FEV1) and SNPs with P-values less than 5×10−8 are considered genome-wide significant. If an intermediate phenotype (eg, pack-years of smoking history) is believed to affect the outcome (eg, FEV1), then a mediation analysis can be run to determine the direct and indirect effects of the SNP on the outcome through the mediator.
Determining which SNPs to include in the mediation analysis is challenging. In GWAS, due to linkage disequilibrium (LD), several SNPs within a gene may be associated with the outcome but these associations are not independent. If every SNP in the region is tested, then this creates a multiple testing issue. If a Bonferroni correction is used, then this would be overly stringent due to the LD in the region. A potential solution is to combine these SNPs that are in LD in the region through aggregate methods such as haplotype analysis or genotype scores. Alternatively, picking the most significant SNP in the region from the GWAS for the outcome may not be appropriate since most mediation methods assume that the SNP chosen is the causal variant. Another approach is to choose the putative causal variant as the SNP of interest for the mediation analysis but this may be difficult in practice. For instance in the CHRNA5/3 region on chromosome 15q25 there is an amino acid substitution (rs16969968, D398N) that alters function of its receptor, which is a plausible candidate for a causal variant in this region. However, there are also SNPs associated with differences in mRNA levels (rs5888765 and rs880395) in LD with rs16969968, which are also putative causal variants. The specific solution to this problem will be influenced by the genetic architecture of the region of interest.
Furthermore, the SNP that is the causal variant for the outcome (eg, FEV1) may not be the causal variant for the mediator (eg, pack-years of smoking history). Picking SNPs for the mediation analysis that are significantly associated with both the mediator and the outcome of interest within one cohort or study will bias the analysis toward an indirect effect in this same cohort or study. Therefore, we recommend only picking SNPs associated with the outcome of interest from the GWAS and not both the mediator and the outcome. Alternatively, SNPs for the mediation analysis may be chosen based on a priori biological reasoning.
Conclusion
Mediation analysis can provide insight into the biologic processes that lead to clinically relevant diseases. As such, evidence from mediation analysis can lead to important therapeutic targets for disease prevention. GWAS are a rich source of evidence for the genetic susceptibility to diseases, but by itself, GWAS do not identify the biological pathways responsible for these associations. Mediation analysis can be an important addition to GWAS when pleiotropy exists at a locus for the disease outcome and the intermediate phenotype by partitioning the observed association with the disease outcome into processes explained by the intermediate phenotype and processes not fully explained by the intermediate phenotype.
GWAS can identify novel genetic associations with disease outcomes. In contrast to candidate gene approaches, GWAS do not assume a specific biologic pathway. As such there is no a priori specific biologic pathway being examined. GWAS significant results may provide insight into the biologic processes that account for the association if the genes have known biological activity. However, these genes may have multiple biologic properties. In addition, GWAS signals may come from genes without known biologic effects or within gene deserts. Regardless, the true biological pathways between the gene and the outcome cannot be known unless formally tested.
Mediation analysis requires a priori assumptions about the direction of the effects observed.19 In addition, the interpretation of “direct” effects may represent a true alternate biologic process, measurement error of the intermediate phenotype, or additional steps along the same biologic pathway. For example, direct effects of CHRNA5/3 on COPD and lung cancer other than through the mediator of pack-years of smoking history may be due to measurement error of self-reported smoking history, additional aspects of smoking not captured by pack-years of smoking such as nicotine dependence, or a true alternate biologic process of either these nicotine receptor genes or other genes in the region that are in LD with CHRNA5/A3.
Combining results from GWAS with mediation analysis provides an important platform for identifying novel genes associated with clinically relevant outcomes and the biologic processes that lead to these associations. This framework may identify therapeutic targets for the treatment of common complex diseases.
Acknowledgments
Research reported in this publication was supported by the National Heart, Lung, and Blood Institute of the National Institutes of Health under Award Number K01HL125858. The content is solely the responsibility of the authors and does not necessarily represent the official views of the National Institutes of Health. This work was also supported by grants R01HL089897 and R01HL089856.
Disclosure
The authors report no conflicts of interest in this work.
References
Chen LS, Hung RJ, Baker T, et al. CHRNA5 risk variant predicts delayed smoking cessation and earlier lung cancer diagnosis-a meta-analysis. J Natl Cancer Inst. 2015;107(5):pii:djv100. | |
Cho MH, McDonald MN, Zhou X, et al. Risk loci for chronic obstructive pulmonary disease: a genome-wide association study and meta-analysis. Lancet Respir Med. 2014;2:214–225. | |
Benowitz NL. Cotinine as a biomarker of environmental tobacco smoke exposure. Epidemiol Rev. 1996;18:188–204. | |
Committee on Passive Smoking, Board of Environmental Studies and Toxicology, National Research Council. Environmental Tobacco Smoke: Measuring Exposures and Assessing Health Effects. Washington, DC: National Academy Press; 1986:1119–1127. Available from: http://www.nap.edu/openbook.php?isbn=0309037301. | |
Siedlinski M, Tingley D, Lipman PJ, et al. Dissecting direct and indirect genetic effects on chronic obstructive pulmonary disease (COPD) susceptibility. Hum Genet. 2013;132:431–441. | |
Lutz SM, Hokanson JE. Genetic influences of smoking and clinical disease: understanding behavioral and biological pathways with mediation analyses. Ann Thorac Med. 2014;11(7):1082–1083. | |
Gu J, Chen M, Shete S, et al. A genome-wide association study identifies a locus on chromosome 14q21 as a predictor of leukocyte telomere length and as a marker of susceptibility for bladder cancer. Cancer Prev Res. 2011;4:514–521. | |
Teng MS, Hsu LA, Wu S, Sun YC, Juan SH, Ko YL. Association of CDH13 genotypes/haplotypes with circulating adiponectin levels, metabolic syndrome, and related metabolic phenotypes: the role of the suppression effect. PLoS One. 2015;10(4):e0122664. | |
Hardy DS, Racette SB, Hoelscher DM. Macronutrient intake as a mediator with FTO to increase body mass index. J Am Coll Nutr. 2014; 33(4):256–266. | |
Sritara C, Thakkinstian A, Ongphiphadhanakul B, et al. Causal relationship between the AHSG gene and BMD through fetuin-A and BMI: multiple mediation analysis. Osteoporos Int. 2014;25:1555–1562. | |
Huang YT, VanderWeele TJ, Lin X. Joint analysis of SNP and gene expression data in genetic association studies of complex diseases. Ann Appl Stat. 2014;8(1):352–376. | |
Huang YT. Integrative modeling of multi-platform genomic data under the framework of mediation analysis. Stat Med. 2015;34:162–178. | |
Pierce BL, Tong L, Chen LS, et al. Mediation analysis demonstrates that trans-eQTLs are often explained by cis-mediation: a genome-wide analysis among 1,800 South Asians. PLoS Genet. 2014;10(12):e1004818. | |
Zhang Q, Feitosa M, Borecki IB. Estimating and testing pleiotropy of single genetic variant for two quantitative traits. Genet Epidemiol. 2014;138(6):523–530. | |
Lutz SM, Vansteelandt S, Lange C. Testing for direct genetic effects using a screening step in family-based association studies. Front Genet. 2013;4:243. | |
Vansteelandt S, Goetgeluk S, Lutz S, et al. On the adjustment for covariates in genetic association analysis: a novel, simple principle to infer direct effects. Genet Epidemiol. 2009;33(5):394–405. | |
Imai, K, Keele L, Tingley D. A general approach to causal mediation analysis, psychological methods. 2010a;15(4):309–334. | |
Imai K, Keele L, Yamamoto T. Identification, inference, and sensitivity analysis for causal mediation effects. Stat Sci. 2010b;25(1):51–71. | |
Vansteelandt S, Lange C. Causation and causal inference for genetic effects. Hum Genet. 2012;131:1665–1676. | |
VanderWeele TJ, Vansteelandt S. Mediation analysis with multiple mediators. Epidemiol Methods. 2013;2:95–115. | |
Baron RM, Kenny DA. The moderator-mediator variable distinction in social psychological research: conceptual, strategic, and statistical considerations. J Pers Soc Psychol. 1986;51:1173–1182. | |
MacKinnon DP, Lockwood CM, Hoffman JM, West SG, Sheets V. A comparison of methods to test mediation and other intervening variable effects. Psychol Methods. 2002;7:83–104. | |
Hayes AF, Preacher KJ. Statistical mediation analysis with a multicategorical independent variable. Br J Math Stat Psychol. 2014;67(3):451–470. | |
Taylor AB, MacKinnon DP. Four applications of permutation methods to testing a single-mediator model. Behav Res Methods. 2012;44:806–844. | |
Sobel ME. Asymptotic confidence intervals for indirect effects in structural equation models. Sociol Methodol. 1982;13:290. | |
Mackinnon DP, Fairchild AJ. Current directions in mediation analysis. Curr Dir Psychol Sci. 2009;18(1):16. | |
VanderWeele TJ, Vansteelandt S. Conceptual issues concerning mediation, interventions and composition. Stat Interface. 2009;2:457–468. | |
VanderWeele TJ, Vansteelandt S. Odds ratios for mediation analysis for a dichotomous outcome. Am J Epidemiol. 2010;172:1339–1348. | |
Hernan MA, Hernandez-Diaz S, Werler MM, Mitchell AA. Causal knowledge as a prerequisite for confounding evaluation: an application to birth defects epidemiology. Am J Epidemiol. 2002;155:176–184. | |
Vansteelandt S. Estimation of direct and indirect effects. In: Berzuini C, Dawid P, Bernardinelli L, editors. Causal Inference: Statistical Perspectives and Applications. Canada: Wiley; 2012. | |
Robins JM, Wasserman L. On the impossibility of inferring causation from association without background knowledge. In: Glymour C, Cooper G, editors. Computation, Causation, and Discovery. Cambridge: AAAI Press/The MIT Press; 1999:305–321. | |
Ten Have TR, Joffe M. A review of causal estimation of effects in mediation analyses. Stat Methods Med Res. 2012;21:77–107. | |
Greenland S. Quantifying biases in causal models: classical confounding vs collider-stratification bias. Epidemiology. 2003;14(3):300–306. | |
Kraft P. Letter to the editor: analyses of genome-wide association scans for additional outcomes. Epidemiology. 2007;18(6):838. | |
Lutz SM, Hokanson JE, Lange C. An alternative hypothesis testing strategy for secondary phenotype data in case-control genetic association studies. Front Genet. 2014;5:188. | |
He J, Li H, Edmondson AC, Rader DJ, Li M. A Gaussian copula approach for the analysis of secondary phenotypes in case control genetic association studies. Biostatistics. 2012;3(3):497–508. | |
Monsees GM, Tamimi RM, Kraft P. Genome-wide association scans for secondary traits using case-control samples. Genet Epidemiol. 2009;33:717–728. | |
Li H, Gail MH. Efficient adaptively weighted analysis of secondary phenotypes in case-control genome-wide association studies. Hum Hered. 2012;73(3):159–173. | |
Lin DY, Zeng D. Proper analysis of secondary phenotype data in case-control association studies. J Genet Epidemiol. 2009;33(3):256–265. | |
Richardson DB, Rzehak P, Klenk J, Weiland SK. Analyses of case control data for additional outcomes. Epidemiology. 2007;18(4):441–445. | |
Wang J, Shete S. Power and type I error results for a bias-correction approach recently shown to provide accurate odds ratios of genetic variants for the secondary phenotypes associated with primary diseases. Genet Epidemiol. 2011;35:739–743. | |
Wang J, Shete S. Estimation of odds ratios of genetic variants for the secondary phenotypes associated with primary diseases. Genet Epidemiol. 2011;35:190–200. | |
Valeri L, Lin X, VanderWeele TJ. Mediation analysis when a continuous mediator is measured with error and the outcome follows a generalized linear model. Stat Med. 2013;33:4875–4890. | |
Ogburn EL, VanderWeele TJ. Analytic results on the bias due to nondifferential misclassification of a binary mediator. Am J Epidemiol. 2012; 176(6):555–561. |
 © 2015 The Author(s). This work is published and licensed by Dove Medical Press Limited. The full terms of this license are available at https://www.dovepress.com/terms.php and incorporate the Creative Commons Attribution - Non Commercial (unported, v3.0) License.
By accessing the work you hereby accept the Terms. Non-commercial uses of the work are permitted without any further permission from Dove Medical Press Limited, provided the work is properly attributed. For permission for commercial use of this work, please see paragraphs 4.2 and 5 of our Terms.
© 2015 The Author(s). This work is published and licensed by Dove Medical Press Limited. The full terms of this license are available at https://www.dovepress.com/terms.php and incorporate the Creative Commons Attribution - Non Commercial (unported, v3.0) License.
By accessing the work you hereby accept the Terms. Non-commercial uses of the work are permitted without any further permission from Dove Medical Press Limited, provided the work is properly attributed. For permission for commercial use of this work, please see paragraphs 4.2 and 5 of our Terms.