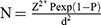Back to Journals » Infection and Drug Resistance » Volume 15
Magnitude and Antimicrobial Susceptibility Profile of Salmonella Recovered from Export Abattoirs Located in East Shewa, Ethiopia
Authors Alemu A, Regassa F, Kebede N, Ambachew R, Girma M, Asefa Z, Tsegaye W
Received 18 November 2021
Accepted for publication 3 March 2022
Published 29 March 2022 Volume 2022:15 Pages 1353—1365
DOI https://doi.org/10.2147/IDR.S348773
Checked for plagiarism Yes
Review by Single anonymous peer review
Peer reviewer comments 3
Editor who approved publication: Professor Suresh Antony
Abayneh Alemu,1,2 Fikru Regassa,2 Nigatu Kebede,3 Rozina Ambachew,1 Musse Girma,3 Zerihun Asefa,4 Wondewosen Tsegaye1
1Department of Medical Microbiology and Parasitology, St. Paul’s Hospital Millennium Medical College, Addis Ababa, Ethiopia; 2Department of Livestock Resource Sector, Ministry of Agriculture, Addis Ababa, Ethiopia; 3Department of Animal Health and Research Unit, Akililu Lemma Institute of Pathobiology, Addis Ababa University, Addis Ababa, Ethiopia; 4Department of Clinical Studies, College of Veterinary Medicine and Agriculture, Addis Ababa University, Bishoftu, Oromia State, Ethiopia
Correspondence: Abayneh Alemu, Addis Ababa, Ethiopia, Tel +251913169301, Email [email protected]
Background: Salmonella is one of the most common foodborne pathogens globally, and it remains a major public health concern with the increasing concern of the emergence and spread of antimicrobial-resistant strains. In Ethiopia, the information on the prevalence of Salmonella is scarce in export abattoirs.
Objective: To estimate the magnitude and antimicrobial susceptibility profile of Salmonella recovered from export abattoirs located in East Shewa, Ethiopia.
Methods: A cross-sectional study was conducted from January to October 2020. In the study, 345 samples were collected from five export abattoirs using a systematic random sampling method. There were 150 carcass swabs (100 from goats and 50 from sheep), 60 goat skin swabs, 60 knife swabs, and 75 human stools. The isolates were identified and characterized using standard bacteriological procedures and confirmed using Salmonella genus-specific primer by polymerase chain reaction. Isolates were subjected to antimicrobial susceptibility to 14 antibiotics using the Kirby–Bauer disk diffusion method, and the results were assessed by using Clinical and Laboratory Standards Institute 2018.
Results: Of the 345 samples, 21 (6.08%; 95% CI 4.9– 11.2%) were positive for Salmonella. The specific prevalence of Salmonella in carcass, skin, and knife swabs were 10 (6.67%; 95% CI 3.5– 11.19%), 7 (11.67%; 95% CI 5.70– 23.00%), and 4 (6.67%; 95% CI 2.50– 16.64%), respectively. There was no statistically significant difference in the occurrence of Salmonella among export abattoirs and types of samples (P> 0.05). In the current study, Salmonella was not isolated from sheep carcass and human stool samples. Among the 21 Salmonella isolates, 7 (33.3%) were resistant to at least 1 of the 14 antimicrobial agents tested and 2 (9.04%) of isolates were resistant to two antibiotics, tetracycline, and streptomycin. All isolates were susceptible to kanamycin, chloramphenicol, cephalothin, gentamycin, and ceftriaxone.
Conclusion: Salmonella was detected in carcass, skin, and knife samples from export abattoirs, which can have serious public health consequences. Some commonly used drugs in veterinary medicine have developed antimicrobial resistance. Therefore, sufficient sanitation at abattoirs, appropriate cooking of carcasses, and rational drug use is strongly advised. Further in-depth study such as serotyping and antimicrobial-resistant gene identification is recommended.
Keywords: antimicrobial susceptibility, carcass, export abattoirs, prevalence, Salmonella
Introduction
Salmonella is a foodborne pathogen that motives morbidity and mortality worldwide.1 Non-typhoidal Salmonella (NTS) enterica subsp. enterica is responsible for causing significant numbers of foodborne diseases in many countries.1 Salmonella is a ubiquitous pathogen disseminated to distinct animals and the environment.2,3 Salmonellosis is one of the predominant foodborne zoonotic and animal husbandry trouble for the duration of the world.4 Globally, about 93.8 million cases of illnesses and 155,000 deaths are associated with gastroenteritis due to Salmonella species every year.5 The World Health Organization (WHO) has measured the global burden of foodborne diseases and estimated that NTS enterica accounted for more than 78 million cases of foodborne illnesses worldwide with about 59,000 deaths in 2010.6 NTS species is one of the most major causes of foodborne disorder and manifested with the aid of diarrhea, bacteremia, and focal suppurative infection.7 Africa was said to have the best burden of foodborne diseases per capita, with a median of 2455 foodborne disability-adjusted life years per 100,000 inhabitants.8 The manner of getting rid of the gastrointestinal tract during slaughtering of animals is considered one of the most essential sources of the carcass, and organ contamination with Salmonella at abattoirs.9 Salmonella contamination in the beef chain can occur at several stages along the food supply chain including production, processing, distribution, retailing, preparing, and handling by the consumer.9
Food contaminated with drug-resistant microorganisms is a most vital hazard to public fitness as antibiotic resistance can switch to other bacteria.10 The emergence and spread of antimicrobial-resistant Salmonella originating from food animals or retail meats have grown to be a serious health hazard worldwide, especially in growing countries.11 The emergence of antibiotic-resistant foodborne pathogens has raised the concern of the public as these pathogens are more virulent, causing an increase in the mortality rate of infected patients.12
Drug resistance occurs as a result of unmonitored use of antibiotics in farms for prophylaxis or as growth promoters.13 This can also signify public health danger by switching of resistant Salmonella strains to humans through consumption of contaminated food and food products. However, the sources and transmission routes of Salmonella in growing countries are poorly understood due to the lack of coordinated-country-wide epidemiological surveillance systems.14
Ethiopia’s export abattoirs are private, state-of-the-art Halal-certified slaughterhouses with livestock reception pens, automatic and semiautomatic mechanical slaughter and processing equipment, chilling rooms, air-conditioned deboning facilities, packaging equipment, freezing facilities, and rendering and effluent treatments. These export abattoirs are licensed by independent Islamic affairs Councils accredited by high-level international bodies that evaluated and provided abattoir standards.15
In Ethiopia, the prevalence of NTS from cattle, carcasses, and hide of slaughtered bovine ranges from 2.75% to 31%, and the incidence of foodborne Salmonella infections has expanded dramatically during the past few years. Studies conducted in different parts of the country have demonstrated the presence of Salmonella in human beings,16,17 and indifferent food animals and food products.16 Several factors including under- and malnutrition, HIV-AIDS, the unhygienic living circumstances, and the close relations between humans and animals may substantially contribute to the occurrence of Salmonellosis.16,17
Assessing the Salmonella in food items through isolation and identification of Salmonella has greater public health importance. Adequately managed and enforced Good Manufacturing Practices and food safety monitoring and surveillance are important components of a modern food supply chain that play a critical role in the control of foodborne pathogens.8 However, food systems in Africa are frequently uncoordinated and poorly regulated, resulting in compromised food safety and protection of public health from foodborne illness.8
In Ethiopia, different studies have been conducted to analyze the prevalence of Salmonella and antimicrobial susceptibility profiles both in veterinary and public health setups.18,19 However, a well-organized epidemiological investigation on magnitude and antimicrobial susceptibility profile of Salmonella from an animal carcass, hide, knife, and human stool is lacking, specifically from export abattoirs. Hence, this study was performed to estimate the magnitude and antimicrobial susceptibility profiles of Salmonella in selected export abattoirs in Ethiopia.
Materials and Methods
Study Areas and Periods
In Ethiopia, export abattoirs including Luna, Organic, Halal, Abyssinia and Aljunia were available in different sites of the country. The study was conducted in three selected areas, namely Modjo, Bishoftu, and Dukem, which are located in the East Shewa Zone of the Oromia region (Figure 1). These areas were selected based on the availability of standardized carcass export abattoirs. The study was carried out from January to October 2020. These export abattoirs work based on rules and regulations of the Ministry of Agriculture. Generally, five export abattoirs are found in Modjo, three in Bishoftu, and one in Dukem. Three abattoirs were selected from Modjo, and one selected from Bishoftu and Dukem. The total number of workers involved in these export abattoirs was 215. The carcass contamination was reduced by performing the criteria of Hazard Analysis Critical Control Points.
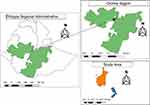 |
Figure 1 Geographical location of the study areas. Notes: |
Study Design
A cross-sectional study design was conducted to estimate the magnitude and antimicrobial susceptibility profile of Salmonella recovered from export abattoirs located in East Shewa, Ethiopia.
Population
Source of population
The source population included goats, sheep, knives, and personnel working in export abattoirs.
Study Population
The study population included apparently healthy, young, male goats, and sheep that were brought for slaughtering, knives, and personnel working in export abattoirs.
Eligibility Criteria
Inclusion Criteria
- Animals that were brought for slaughtering and that did not receive antibiotics treatment.
- A carcass handler with no clinical symptoms of infection and who did not receive antibiotics treatment were included for the human subject part study.
Exclusion Criteria
- Sick animals that were already being treated with antibiotics and people with clinical symptoms (eg, headache, sneezing, and discomfort).
Operational Definition
Multidrug-Resistance
Resistant to three or greater antimicrobial classes.12
Export Abattoir
This is a slaughter facility in respect of which a registration certificate has been issued for the purpose of exporting meat and meat products.
Sample Size Determination
The sample size was calculated by applying the single population proportion estimation formula with a 95% confidence level and with a margin error of 5% desired precision.
The sample size was calculated based on 5.7% and 3.57% expected prevalence of bovine and ovine samples respectively from a previous study done in Addis Abeba Abattoirs Enterprise, Ethiopia.2
The sample size was calculated using the following equation:
where N = required sample size, Z = standard normal deviation (1.96) at 95% confidence level, Pexp = expected prevalence, and d = desired absolute solution (0.05).
Accordingly, the calculated sample size was 150 considering a 10% non-response rate. However, a total of 345 samples (150 carcass swabs [100 from goats and 50 from sheep], 60 goats skin swabs, 60 knife swabs, and 75 human stool samples) were collected for detection of Salmonella. Based on the annual slaughtering capacity of sheep and goats, as well as the number of personnel working in each abattoir, the calculated sample size was proportionally distributed to the five export abattoirs.
Sampling Techniques
To recruit the study participants, a systemic random sampling method was used to enroll eligible study participants. The numbers of study participants to be enrolled from each selected export abattoir were determined by proportionality (based on animal and human study participant load).
Data Collection Procedure
Socio-Demographic Characteristics
For the human sample, after obtaining written consent from all study participants, a semi-structured questionnaire was used to collect socio-demographic characteristics.
Sample Collection and Transportation
Carcass Swabs
Samples were collected from the carcass (n=150, 100 from goats, and 50 from sheep). Each carcass used to be sampled from four regions: neck, abdomen, thorax, and breast. Sampling areas were delineated via sterile aluminum foil templates (10×10 cm) resulting from a total area of 400 cm2. A sterile cotton-tipped swab (2×3 cm) was first soaked in 9 mL of buffered peptone water (BPW) (Oxoid, England) and rubbed over delineated area horizontally and vertically.2 Two sterile cotton-tipped swabs were used to acquire sampling areas.
Skin Swabs
Samples were collected from external goat’s skin surfaces (n=60). Four regions were selected to collect the skin swabs: abdomen, thorax, neck, and breast. A sterile aluminum foil template (10×10 cm) resulted in a total area of 400 cm2 placed in these regions. A sterile cotton-tipped swab with wooden shaft was first soaked in 9 mL of sterile BPW and rubbed over delineated area horizontally and vertically.9 Two sterile cotton-tipped swabs were used to collect the consecutive regions.
Knife Swabs
Samples from the knives (n=60) were collected aseptically using sterile cotton swabs. It was done by rubbing both sides of the knife using a pre-soaked swab.11 Swab samples were collected from the animals selected for slaughtering.
Stool Samples
After proper instruction, each study participant was informed to bring freshly voided stool in a clean, dry, and leak-proof disposable stool cup, and a total of 75 stool specimens were collected.
Upon completion of all swabbing processes, the wooden shaft was broken off and the cotton swab was left interior the test tubes containing 9 mL of sterilized BPW. The swab samples within tubes were shaken for 30 seconds for uniform distribution of microorganisms earlier than transportation. All samples were labeled, positioned in separate plastic bags, transferred into a sterile icebox, and transported to Medical Microbiology Laboratory, Aklilu Lemma Institute of Pathobiology (ALIPB) within 3–4 hours for the isolation of Salmonella.
Microbiological Analysis
Isolation and Identification of Salmonella
Isolation and identification of Salmonella have been performed at the Department of the Medical Microbiology Laboratory of ALIPB. Salmonella isolation and identification had been carried out in line with International Organization for Standardization (ISO), ISO 6579:2002/Amd 1:2007, and global Salmonella surveillance and laboratory support of the World Health Organization: Laboratory Protocols (Identification of Salmonella).20,21
Pre-Enrichment in Nonselective Liquid Medium
The swab samples were soaked in 9 mL of BPW. This pre-enriched sample incubated for 18–24 hours at 37°C for recovery and proliferation of cell would possibly be injured during processing or to make a number of the target organism grow to detectable level.4
Enrichment Selective Liquid Media
Enrichment selective broths, namely Rappaport Vassiliadis Soya (RVS) (Oxoid, England CM950-500G) for all samples except stools and Selenite F for a stool sample (Oxoid, England, CM651-500G), were used to inhibit nontargeted microorganisms like gram-positive bacteria and coliforms and approve fast multiplication of Salmonella. After pre-enrichment in 0.1 mL of culture from BPW were transferred aseptically into 10 mL of RVS Broth and incubated for 18–24 hours at 41.5°C. For a stool sample, 0.1 mL of cultures from BPW was transferred to 10 mL of Selenite F Broth, homogenized, and incubated for 18–24 hours at 37°C.19
Plating Out and Identification
A loopful of 100 µm used to be taken from RVS and Selenite F Broths, streaked into the xylose lysine deoxycholate (XLD) agar (Oxoid, England, CM0469-500 G) plates, and incubated for 18–24 hours at 37°C. Suspected Salmonella isolates were subcultured on nutrient agar (Oxoid, England, CM0003-500G) and incubated for 18–24 hours at 37°C. The isolates from the subculture were stored in the refrigerator at 4°C for the biochemical test, molecular testing, and antimicrobial susceptibility test.4
Biochemical Characterization
Suspected Salmonella colonies from nutrient agar (Oxoid, England CM0003-500G) were picked up and its biochemical characteristics were determined using triple sugar iron agar (Oxoid, England CM277-500G), lysine iron agar (Oxoid, England CM0381-500G), Simmon’s citrate agar (Himedia, India CM0129-500G), urea slant (Himedia, India M111A-500G), and sulfide indole motility agar (Oxoid, England S12-500G).3
Molecular Techniques
Polymerase chain reaction (PCR) was used to confirm the identification made by phenotypic tests.
DNA Extraction
Bacterial colonies confirmed as Salmonella with the aid of biochemical tests were cultured overnight on XLD agar. Then DNA extraction was carried out using the boiling method. The set of primer targeted conserved regions of Salmonella forward and reverse were used.13
Molecular Confirmation by Using a PCR
All isolates that showed specific biochemical characteristics of Salmonella were further confirmed via the use of a genus-specific PCT.1 It is based on the amplification of a 496-base pair (bp) segment of histidine transport operon gene (Table 1), which is noticeably conserved among species of Salmonella. Reference strain of S. typhimurium (ATCC 14028) was used as positive control during PCR. PCR amplification was once run in reaction mixtures (20 μL) with master mix (10 μL), forward (0.50 μL) and reverse (0.50 μL) primer, nuclease-free water (8.0 μL), and DNA template (1.0 μL). Amplification was carried out in a thermocycler with temperature profiles of 2 minutes at 94°C for initial denaturation observed through 35 cycles of at 94°C for 1 minute, annealing at 58°C for 1 minute, and extension at 72 °C for 1 minute with remaining extension step at 72°C for 5 minutes.1
 |
Table 1 Primers Used to Detect Salmonella |
Agarose Gel Electrophoresis and Visualization of PCR Products
PCR products were electrophoresed using 2 grams agarose powder (Rugby, UK) in 100 mL of 1× TAE buffer (Bio Concept, Switzerland). A quantity of 2 µL of ethidium bromide was mixed with the gel before pouring it into the casting tray. A 100-bp DNA ladder was used as a molecular dimension marker to estimate the size of the products. A band of 496 bp was regarded as superb for Salmonella. Gel electrophoresis used to be carried out at 120 volts for 60 minutes considered beneath under an ultraviolet trans-illuminator.13
Antimicrobial Susceptibility Test
The isolates confirmed by PCR had been subjected to antibiotic susceptibility test using Kirby–Bauer disk diffusion techniques,22 on Mueller–Hinton agar (MHA; Oxoid, England), and results interpreted according to Clinical and Laboratory Standards Institute (CLSI, 2018).23 Antibiotics have been selected based on the routine prescription for human and animal use and their availability on the market of the country. From each PCR confirmed isolate, three to four colonies grown on nutrient agar were transferred to a tube containing 3 mL of nutrient broth (Oxoid, England). The broth culture was incubated for 18–24 hours at 37°C untill its turbidity was adjusted to 0.5 McFarland standards. The suspension was once inoculated onto MHA plates using sterile cotton swabs. The plates have been uniformly inoculated via rubbing toward the whole agar surface and rotating the plates thrice at about 90° degrees. The plates were held at room temperature for 15 minutes to allow drying. Antibiotic-impregnated disks were applied to the floor of the inoculated plate using sterile forceps and incubated aerobically for 24 hours at 37°C. Salmonella isolates examined for the antibiotics include kanamycin (30 μg, K), ciprofloxacin (5 μg, CIP), chloramphenicol (30 μg, C), cephalothin (30 μg, CEP), tetracycline (30 μg, TE), ampicillin (10 µg, AMP), nalidixic acid (30 µg, NA), streptomycin (10 μg, S), gentamicin (10 µg, GEN), ceftriaxone (30 μg, CRO), amikacin (10 μg, AN), neomycin (30 μg, N), amoxicillin+clavunic acid (20/10 μg, AMC), and sulfamethoxazole+trimethoprim (1.25/23.75 µg, SXT). Following the incubation of the plates for 24 hours at 37°C, the diameter of the inhibition zone was measured to the nearest millimeter using a digital caliper and interpreted as sensitive (S), intermediate (I), or resistant (R) in accordance with CLSI 2018.23
Quality Controls
Standard operating procedures of the laboratory ensured the reliability and validity of test results. A batch of the media was incubated for 24 hours at 37°C, and the media were checked for pH, sterility, ability, and support growth before use. In addition to these, visually the media were checked for depth, smoothness, hemolysis, excessive bubbles, contamination, check for cracked or damaged plates, and frozen or melted agar before use. The media performances were checked with a known positive control standard American Type Culture Collection (ATCC) reference: S. typhimurium ATCC 14028 as positive control and E. coli ATCC 25922 as negative control.
Data Analysis
Data were entered and analyzed using STATA version 14. Fisher’s exact test was used to investigate significant differences among abattoirs and kinds of samples. P-value <0.05 was considered as indicative of a statistical significance difference.
Results
Socio-Demographic Characteristics of Human Study Participants in Selected Export Abattoirs
A total of 75 carcass handlers were enrolled in this study with a response rate of 100%. The sex distributions were 55 men and 20 women with an age range of 22–50 years with a mean age of 32 years (± 8.61 SD). Educational backgrounds were as follows: 3 (5%) primary school, 65 (86.6%) secondary school, and 7 (9.33%) diploma. Most of the respondents were married (50 (67%)). All participants were trained about food hygiene and wearing personnel protective equipment. Among carcass handlers, 65 (86.6%) have certificates in food safety. All participants undertook periodical medical check-ups and washed their hands with soap and water after using the toilet or touching any material (Table 2).
 |
Table 2 Socio-Demographic Characteristics of Human Study Participants in Selected Export Abattoirs, East Shewa, Ethiopia, from January to October 2020 |
Prevalence of Salmonella
During the study period, a total of 345 samples were collected from five export abattoirs. Thirty-six samples showed suspected Salmonella colonies on XLD agar and only 21 (6.08%; 95% CI 4.9–11.2%) isolates showed typical biochemical properties of Salmonella. These 21 isolates were further confirmed by PCR amplification (Figure 2). Salmonella was not isolated from all human stool and sheep carcass swabs. The highest proportion of positivity was detected from goat skin swabs 7/60 (11.67%) followed by goat carcass swabs 10/150 (6.67%), and knife swabs 4/60 (6.67%) (Table 3).
 |
Table 3 Detection Rate of Salmonella on Different Samples in Selected Export Abattoirs, East Shewa, Ethiopia, from January to October 2020 |
Antimicrobial Susceptibility Testing of Salmonella Isolates
Among the 21 molecular confirmed Salmonella isolates, all isolates were susceptible to kanamycin, chloramphenicol, cephalothin, gentamycin, and ceftriaxone. A total of 21 (100%) and 1 (4.76%) isolates were intermediately resistant to neomycin, and streptomycin respectively (Table 4).
 |
Table 4 Antibiotic Susceptibility Profiles of Salmonella Isolates (n=21) in Selected Export Abattoirs, East Shewa, Ethiopia, from January to October 2020 |
Overall, 7 (33.33%) of the isolates were resistant to at least 1 of the tested 14 antimicrobial agents. Among the 21 isolates, only 2 (9.04%) were resistant to tetracycline and streptomycin (Table 5).
 |
Table 5 Drug-Resistance Patterns of Salmonella Isolates from Studied Ethiopian Export Abattoirs from January to October 2020 |
Discussion
Foodborne gastroenteritis caused by NTS represents a major public health problem worldwide. As Salmonella is transmitted through contaminated food or water, its presence in food animals and animal products has relevant public health implications. Thus, monitoring food safety is a key point in preventing and controlling the spread of Salmonella, as well as in providing healthier food products.3 In this study, the overall prevalence of Salmonella was 21 (6.08%) with 95% CI 4.90–11.2%.. From this, 10 (6.67%), 7 (11.67%), and 4 (6.67%) were from goat carcass swabs, goat skin swabs, and knife swabs, respectively. The prevalence of Salmonella among different samples in this study showed no statistically significant association (P>0.05).
The finding of the current study should be comparable with a study reported in dairy cattle in central Ethiopia (7%),24 Colorado State University veterinary teaching hospitals (5.9%),25 and on pork and goat carcass in the Bahamas (5.9%)26 On the contrary, our finding is lower than the study reported on exotic chickens in Debre Zeit and Modjo, Ethiopia (14.6%)27; ground beef at a retail store in Jalisco State, Mexico (56.7%)28; from milk and meat in Bangladesh (60%),29 Kwata slaughterhouse Awka, Anambra State (33.5%)16; from abattoir and environment in Nigeria (92.31%)30; and raw beef in Wolaita Sodo municipal abattoir, Southern Ethiopia (12.5%).31 However, the current result is higher than the study conducted on slaughtered cattle in Addis Ababa, Ethiopia (3.7%)3; slaughter sheep in Turkey (0.7%)32; food handlers at the University of Gondar, Ethiopia, that suggested (3.1%)33; from animal-origin food items in Gondar, Ethiopia (5.5%)34; and slaughtered bovine and ovine in Addis Ababa Abattoirs Enterprise (4.64%).2 Salmonella was not detected from sheep carcass swabs, which is comparable with the study conducted on slaughter sheep carcass swab in Turkey.32 The authors would like to factor out that the prevalence may be underestimated because the sample in the current study was a swab rather than meat, and the protocol used to be not the parallel ISO method, which is more sensitive to detect specific serovars. The discrepancies between our study and others could be associated with the degree of exposure of animals to stress factors like transportation and starvation, climatic conditions, management practices, age-groups, species of animals, hygienic conditions adopted kinds of abattoirs and facilities, food coping with and geographical difference.
In the current study, the proportion of Salmonella was isolated from skin swabs (11.66%), carcass swabs (6.66%), and knife swabs (6.66%). This result is relatively lower in proportion study conducted to the cattle slaughtered in South Africa for Salmonella isolation suggested carcass swabs (30%), skin swabs (59.7%),4 and knife swabs (16.7%) selected from dairy farms, abattoir, and humans at Asella Town, Ethiopia.9 However, it is higher than the study conducted on carcass swabs on animal sources in South Africa which reported (1.8%),35 knife swabs (2.5%) from abattoir and environment in Nigeria,30 and (1.6%) skin swabs of dairy cattle slaughterhouse in Northern Italy,36 and (4.5%) Kwata slaughterhouse, Awka, Anambra State.16 This difference may be attributed to hygienic status, management systems, and cross-contamination among materials used in slaughtering procedures.
Animals that entered abattoirs have been particularly dirty, which contributed to the spread and cross-contamination of skin with the Salmonella pathogen. The proportion of carcass contamination recorded in this study, as well as the potential sources of contamination, are diverse. Contact between aprons and then the carcass is unavoidable in many areas and may also result in the carcass-to-carcass transfer of Salmonella.37 The presence of the Salmonella execrations in batches of animals in transit and passing through the lairage should result in contamination of the skin.37 Moreover, it indicated that the exterior surface of the animals serves as a source of the illness for the underlying sterile carcass floor in the course of the dehiding process.38 It has been indicated that manual operation of all the processing steps in the slaughtering of the animals in the abattoirs, as a substitute than the use of semiautomatic or automatic system in operation will increase the chances of contamination of edible organs and spreading of Salmonella in abattoirs’ environment.39 Occasionally, when transferring carcass from one area to another on the floor; there is close contact between the different carcass, and this may also result in the transfer of Salmonella carcass-to-carcass as well as leftovers anal surfaces feces to the carcass. All these elements may also play a large role in the occurrence of Salmonella in chosen export abattoirs in East Shewa, Ethiopia.
In this study, we also assessed the antimicrobial susceptibility of Salmonella isolates. The result of the in vitro antibiotics’ sensitivity test to Salmonella isolates showed distinct degrees of sensitivity toward tested antibiotics ranging from 0% to 100%. The highest susceptibility (100%) was determined toward kanamycin, chloramphenicol, cephalothin, gentamycin, and ceftriaxone while some resistance to tetracycline and streptomycin were seen. Tetracycline and streptomycin have shown greater resistance rates, which could be attributed to the fact that they are among the most commonly used antimicrobials for treating several infectious diseases in livestock. The isolates were susceptible to ceftriaxone and chloramphenicol, which is in settlement with the preceding of cattle slaughtered in Addis Abeba,3 and dairy cattle in central Ethiopia.24 In addition, isolates in the current study were susceptible to gentamycin and ceftriaxone, which is in line with preceding studies from food handlers at the University of Gondar, Ethiopia.33
Among the 21 Salmonella isolates, 7 (33.3%) were resistant to at least one antimicrobial agent and 2 (9.04%) of Salmonella isolates were resistant to tetracycline and streptomycin. This conforms with the study conducted in Ecuador that reported resistance rate for other antibiotics ranged from 11.1% up to 33.3%.40 Antibiotic resistance is the evolutionary response by bacteria to strong selective pressure that results from exposure to antibiotics.13 This could be an indicator of the acquisition of the resistance genes for those drugs due to the indiscriminate use of drugs at subtherapeutic doses in feed additives to promote growth creating an on-farm selection of antimicrobial-resistant strains. Hence, continued misuse of antimicrobials may exert stress on resistant bacteria, favoring their emergence and spread. The low prevalence of tetracycline resistance discovered in this study was recorded. This is due to the fact that younger animals are preferred for slaughter, particularly in export abattoirs. When an animal is young, it has a lower threat of acquiring an antimicrobial throughout its continue to be in the community. Antimicrobial resistance is a global public health problem.4 Resistance to antimicrobials ought to be due to three basic mechanisms: 1) modification of the antibiotic by decreasing absorption or increasing efflux of the antibiotic by using their enzymes, 2) change in the target site of the antibiotic, and 3) acquisition of the ability to break or modify the antibiotic.41 Several lines of evidence demonstrate that the use of antimicrobial agents in food animals contributes to the emergence and dissemination of antimicrobial resistance in foodborne Salmonella.4 Multidrug resistance has not been recorded in this study. In this study, among the tested antibiotics, kanamycin, ceftriaxone, chloramphenicol cephalothin, and gentamicin were found to be the most effective drugs to inhibit the in vitro growth of these isolates. Thus, these drugs could be used for empirical therapy in the areas where culture facility is no longer available.
Limitation of the Study
In this study, isolates were not serotyped or molecularly characterized due to financial constraints. Selection bias can also be viewed as a constraint.
Conclusion and Recommendation
Investigating the incidence and antimicrobial susceptibility of Salmonella from an animal carcass, skin, knife swabs, and humans in export abattoirs is of paramount significance to techniques of minimizing the possible transmission of Salmonella between people and animals. Moreover, it is vital in combating the emergence of antibiotic-resistant of Salmonella. The information gathered in this cross-sectional study, collectively with different comparable studies, is important to acquire the importance of studying Salmonella in export abattoirs. In general, from this study, it can be concluded that the prevalence of Salmonella is 21 (6.08%), which appears to be high. This result is significantly high to be a potential source of foodborne salmonellosis. Thus, bacteriological evaluation of Salmonella pathogen from export abattoirs was crucial to improving the surveillance system and hygienic standards.
Among the tested antimicrobials, kanamycin, ceftriaxone, chloramphenicol, cephalothin, and gentamicin were 100% susceptible. Antimicrobial treatment techniques need to be based on bacteriological culture followed by antimicrobial susceptibility tests.
Abbreviations
ALIPB, Aklilu Lemma Institute of Pathobiology; BPW, buffered peptone water; CLSI, Clinical and Laboratory Standards Institute; PCR, polymerase chain reaction; RVS Broth, Rappaport Vassiliadis Soya Broth; SPHMMC, Saint Paul’s Hospital Millennium Medical College; WHO, World Health Organization; XLD, xylose lysine deoxycholate.
Data Sharing Statement
Data is available upon request from the corresponding author.
Ethical Approval and Consent to Participate
Ethical approval was obtained from St. Paul’s Hospital Millennium Medical College (SPHMMC) (Pm23/423) Institutional Review Board (IRB). Written informed consent was obtained from the study participant before the initiation of data collection and it was performed in accordance with the Declaration of Helsinki, and abattoir owners were informed and aware of the purpose of the study. The personal results of any investigation remained confidential. All recognized cases of Salmonella in export abattoirs were referred to attending veterinary supervisors. The best practice of veterinary care was taken in sampling part of the animals.
Acknowledgments
This study was once financially supported by the Ministry of Agriculture, Livestock Resource Sector, Akililu Lemma Institute of Pathobiology, Addis Ababa University, and St. Paul’s Hospital Millennium Medical College. We additionally extend our profound gratitude to study participants for their willingness without whom this research work would not have been possible.
Author Contributions
All authors made a significant contribution to the work reported, whether that is in the conception, study design, execution, acquisition of data, analysis, and interpretation, or in all these areas; took part in drafting, revising, or critically reviewing the article; gave final approval of the version to be published; have agreed on the journal to which the article has been submitted; and agree to be accountable for all aspects of the work.
Funding
This study was sponsored by the Ministry of Agriculture, Livestock Resource Sector, and St. Paul’s Hospital Millennium Medical College, Addis Ababa, Ethiopia, which furnished full financial support.
Disclosure
The authors declare that there is no conflicts of interest regarding the publication of this paper.
References
1. Madoroba E, Kapeta D, Gelaw AK. Salmonella serovars and antimicrobial resistance profiles of cattle slaughtered in South Africa. J Vet Res. 2014;83(1):1–8.
2. Kebede A, Kemal J, Alemayehu H, Mariam SH. Isolation, identification and antibiotic susceptibility of Salmonella from slaughtered bovines and ovines in Addis Ababa abattoir enterprise, Ethiopia: cross-sectional study. Int J Bacteriol. 2016;3(10):1–8. doi:10.1155/2016/3714785
3. Ketema L, Ketema Z, Kiflu B, et al. Prevalence and antimicrobial susceptibility profiles of Salmonella serovars isolated from slaughtered cattle in Addis Ababa, Ethiopia. Biomed Res Int. 2018;34(10):1–7. doi:10.1155/2018/9794869
4. Ali DA, Tadesse B, Ebabu A. Prevalence and antibiotic resistance pattern of Salmonella isolated from caecal contents of exotic chicken in Debre Zeit and Modjo, Ethiopia. Hindawi. 2020;2020(10):1–6.
5. Geresu MA, Desta WZ. Carriage, risk factors, and antimicrobial resistance patterns of Salmonella isolates from raw beef in Jimma, South Western Ethiopia. Infect Drug Resist. 2021;14:2349–2360. doi:10.2147/IDR.S313485
6. Realpe-quintero M, Barba-león J, Perez-montaño JA, Cabrera-diaz E. Genetic diversity and antimicrobial resistance of Salmonella serotypes recovered throughout the beef production chain and from patients with salmonellosis. Peer J. 2018;6:1–20. doi:10.7717/peerj.5482
7. Arguello H, Álvarez-ordonez A, Carvajal A, Rubio P, Prieto M, Rubio P, Prieto M. Role of slaughtering in Salmonella spreading and control in pork production.J Food Prot. 2013;76(5):899–911. doi:10.4315/0362-028X.JFP-12-404
8. Cummings KJ, Warnick LD, Alexander KA, et al. The duration of fecal Salmonella shedding following clinical disease among dairy cattle in the northeastern USA. Prev Vet Med. 2009;92(1):134–139. doi:10.1016/j.prevetmed.2009.07.002
9. Beyene T, Yibeltie H, Chebo B, et al. Identification and antimicrobial susceptibility profile of Salmonella isolated from selected dairy farms, abattoir, and humans at Asella Town, Ethiopia. J Veterinary Sci Techno. 2016;7(3):1–7.
10. Akbar A, Anal AK. Isolation of Salmonella from ready-to-eat poultry meat and evaluation of its survival at low temperature, microwaving and simulated gastric fluids. J Food Sci Technol. 2015;55(5):3051–3057. doi:10.1007/s13197-014-1354-2
11. Thai TH, Hirai T, Lan NT, Shimada A, Ngoc PT. Antimicrobial resistance of Salmonella serovars isolated from beef at retail markets in the North Vietnam. Bacteriol Antimicrobial. 2012;12(53):1163–116.9.
12. Eng S, Pusparajah P, Mutalib NA, Ser H, Chan K, Lee L. Salmonella: a review on pathogenesis, epidemiology and antibiotic resistance. Front Life Sci ISSN. 2015;8(3):284–293. doi:10.1080/21553769.2015.1051243
13. Nyabundi D, Onkoba N, Kimathi R, et al. Molecular characterization and antibiotic resistance profiles of Salmonella isolated from fecal matter of domestic animals and animal products in Nairobi. Trop Dis Travel Med Vaccines. 2017;3(2):1–7. doi:10.1186/s40794-016-0045-6
14. Kaferstein F. Foodborne diseases in developing countries: etiology, epidemiology, and strategies for prevention. Int J Environ Health Res. 2003;13(1):161–168. doi:10.1080/0960312031000102949
15. Ethiopian Meat Producer-Exporters Association. Available from: www.empea.com.et.
16. Nyeleti C, Hildebrandt G, Kleer J, Molla B. Prevalence of Salmonella in Ethiopian cattle and minced beef. Berliner und Münchener tierarztliche Wochenschrift. 2000;113:431–434.
17. Tadesse G, Gebremedhin EZ. Prevalence of Salmonella in raw animal products in Ethiopia: a meta-analysis. BMC Res Notes. 2015;8(163):1–8. doi:10.1186/s13104-015-1127-7
18. Tesfaye W, Melese A, Henok S, Yohanis M. Prevalence and antimicrobial susceptibility profile of Salmonella species from ready-to-eat foods from catering establishments in Jigjiga City, Ethiopia. African J Microbiol Res. 2016;10(37):1555–1560. doi:10.5897/AJMR2016.7944
19. Addis Z, Kebede N, Sisay Z, et al. Prevalence and antimicrobial resistance of Salmonella isolated from lactating cows and contact humans in dairy farms of Addis Ababa: a cross-sectional study. BMC Infect Dis. 2011;11(222):1–19. doi:10.1186/1471-2334-11-222
20. International Organization for Standardization (ISO). ISO 6579:2002/Amd 1:2007. Detection of Salmonella spp. in animal faeces and in environmental samples from the primary production 1 Annex D. In. Microbiology of Food and Animal Feeding Staffs Horizontal Method for the Detection for the Detection of Salmonella spp. Geneva Switzerland: International Organization for Standardization; 2007.
21. Hendriksen RS. A global Salmonella surveillance and laboratory support of the World Health Organization: Laboratory Protocols (Identification of Salmonella). Foodborne Pathog Dis. 2003;8(8):887–900.
22. Hudzicki J. Kirby-Bauer disk diffusion susceptibility test protocol author information. Am Soc Microbiol. 2009;2012:1–23.
23. Performance Standard for Antimicrobial Susceptibility Testing.
24. Eguale T, Engidawork E, Gebreyes WAD, et al. Fecal prevalence, serotype distribution and antimicrobial resistance of Salmonella in dairy cattle in central Ethiopia. BMC Microbiol. 2016;16(20):1–11. doi:10.1186/s12866-016-0638-2
25. Burgess BA, Morley PS. Risk factors for shedding of Salmonella enterica among hospitalized large animals over 10 years period in a veterinary teaching hospital, Colorado State University. J Vet Intern Med. 2019;10(33):2239–2248. doi:10.1111/jvim.15579
26. Hanlon KH, Echeverry A, Miller MF, Brashears MM. Establishment of preliminary baseline of Salmonella presence on pork, and goat carcasses harvested in the Bahamas to address food and nutritional security interventions. Am Soc Anim Sci. 2018;8(4):1–7.
27. Asfaw D, Tadesse B, Ebabu A. Prevalence and antibiotic-resistance patterns of Salmonella isolated from caecal contents of exotic chicken in Debre Zeit and Modjo, Ethiopia. Int J Microbiol. 2020;6(10):1–6. doi:10.1155/2020/1910630
28. Gonzalez D, Barba J, Pacheco C, et al. Frequency, and antimicrobial resistance of Salmonella serotypes on beef carcasses at small abattoirs in Jalisco State, Mexico. Food Prot. 2018;75(5):867–873.
29. Rahman MA, Rahman AK. Detection of multidrug-resistant Salmonella from milk and meat in Bangladish. Bangl J Vet Med. 2016;16(1):115–120. doi:10.3329/bjvm.v16i1.37388
30. Igbinosa EO, Beshiru A. Isolation, and characterization of antibiotic susceptibility profile of Salmonella species isolated from abattoir, and environment. J Sci. 2017;19(2):389–397.
31. Wabeto W, Abraham Y, Anjulo A. Detection and identification of antimicrobial resistant Salmonella in raw beef at Wolaita Sodo municipal abattoir, Southern Ethiopia. J Heal Popul Nutr. 2017;36(52):1–7.
32. Cetin E, Temelli S, Eyigor A. Non-typhoid Salmonella prevalence, serovar distribution and antimicrobial resistance in slaughter sheep, Turkey. Food Sci Anim Resour. 2020;40(1):21–33. doi:10.5851/kosfa.2019.e75
33. Garedew L, Wondafrash N, Feleke A. Identification of drug-resistant Salmonella from food handlers at the University of Gondar, Ethiopia. BMC Res Notes. 2016;7(545):1–9.
34. Ejo M, Garedew L, Alebachew Z, Worku W. Prevalence and antimicrobial resistance of Salmonella isolated animal-origin food items in Gondar, Ethiopia. Biomed Res Int. 2016;10(10):1–8. doi:10.1155/2016/4290506
35. Gelaw AK, Nthaba P, Matle I. Detection of Salmonella from animal sources in South Africa between 2007 and 2014. J S Afr Vet Assoc. 2018;89:1–10. doi:10.4102/jsava.v89i0.1643
36. Bonardi S, Bruini I, Magnani R, Cannistrà N, Brindani F. Low prevalence of Salmonella enterica in dairy cattle at slaughter in Northern Italy. Ital J Food Saf. 2017;6(6172):1–4.
37. Akafete T, Haileleul N. Assessment of risk factors and prevalence of Salmonella in slaughtered small ruminants and environment in an export abattoir, Modjo, Ethiopia. Am-Euras J Agric En Viron Sci. 2011;10(6):992–999.
38. Nabbut NH, Ali-Nakli HM. Incidence of Salmonella in lymph nodes, spleen and feaces of sheep and goats slaughtered in Riyadh public abattoir. J Food Prod. 1982;45:1314–1317. doi:10.4315/0362-028X-45.14.1314
39. Vinueza-Burgos C, Cevallos M, Ron-Garrido L, Bertrand S, De Zutter L. Prevalence and diversity of Salmonella serotypes in Ecuadorian broilers at slaughter age. Plos One. 2016;11(7):1–12. doi:10.1371/journal.pone.0159567
40. Tesfaye H, Alemayehu H, Desta AF, Eguale T. Antimicrobial susceptibility profile of selected Enterobacteriaceae in waste water samples from health facilities, abattoir, downstream rivers and a WWTP in Addis. Antimicrob Resist Infect Control. 2019;8(134):1–11. doi:10.1186/s13756-019-0588-1
41. Aragaw K, Molla B, Muckle A. The characterization of Salmonella serovars isolated from apparently healthy slaugh tered pigs at Addis Ababa abattoir, Ethiopia. Prev Vet Med. 2007;82(3–4):252–261. doi:10.1016/j.prevetmed.2007.05.022
 © 2022 The Author(s). This work is published and licensed by Dove Medical Press Limited. The full terms of this license are available at https://www.dovepress.com/terms.php and incorporate the Creative Commons Attribution - Non Commercial (unported, v3.0) License.
By accessing the work you hereby accept the Terms. Non-commercial uses of the work are permitted without any further permission from Dove Medical Press Limited, provided the work is properly attributed. For permission for commercial use of this work, please see paragraphs 4.2 and 5 of our Terms.
© 2022 The Author(s). This work is published and licensed by Dove Medical Press Limited. The full terms of this license are available at https://www.dovepress.com/terms.php and incorporate the Creative Commons Attribution - Non Commercial (unported, v3.0) License.
By accessing the work you hereby accept the Terms. Non-commercial uses of the work are permitted without any further permission from Dove Medical Press Limited, provided the work is properly attributed. For permission for commercial use of this work, please see paragraphs 4.2 and 5 of our Terms.