Back to Journals » Cancer Management and Research » Volume 12
MAGE-A11 Expression Predicts Patient Prognosis in Head and Neck Squamous Cell Carcinoma
Authors Jia S , Zhang M , Li Y, Zhang L, Dai W
Received 8 November 2019
Accepted for publication 11 February 2020
Published 26 February 2020 Volume 2020:12 Pages 1427—1435
DOI https://doi.org/10.2147/CMAR.S237867
Checked for plagiarism Yes
Review by Single anonymous peer review
Peer reviewer comments 3
Editor who approved publication: Dr Eileen O'Reilly
Shiheng Jia, 1–4 Minghui Zhang, 1–4 Yanshu Li, 1–3 Lan Zhang, 5 Wei Dai 5
1Key Laboratory of Cell Biology, Ministry of Public Health, China Medical University, Liaoning, Shenyang 110122, People’s Republic of China; 2Key Laboratory of Medical Cell Biology, Ministry of Education, China Medical University, Liaoning, Shenyang 110122, People’s Republic of China; 3Department of Cell Biology, China Medical University, Liaoning, Shenyang 110122, People’s Republic of China; 4Department of Clinical Medicine, China Medical University, Liaoning, Shenyang 110122, People’s Republic of China; 5Department of Oral and Maxillofacial Surgery, School and Hospital of Stomatology, China Medical University, Liaoning Provincial Key Laboratory of Oral Diseases, Liaoning, Shenyang 110002, People’s Republic of China
Correspondence: Wei Dai
Department of Oral and Maxillofacial Surgery, School and Hospital of Stomatology, China Medical University, Liaoning Provincial Key Laboratory of Oral Diseases, Liaoning, Shenyang 110002, People’s Republic of China
Tel +86-139-0988-4820
Email [email protected]
Background: Head and neck squamous cell carcinomas (HNSCCs) are the sixth most common cancer worldwide. Growing evidence showed that Melanoma-associated antigen-A11 (MAGE-A11) was abnormally expressed in various malignancies, but MAGE-A11 expression and its biological roles in HNSCC had not been reported in detail. The aim of the study was to investigate the association between MAGE-A11 signatures and clinicopathological features of HNSCC patients and uncover its potential mechanisms in HNSCC patients.
Methods: In the present study, we analyzed the expression of MAGE-A11 gene and evaluated the impact of MAGE-A11 genes expression on clinical outcome from the Cancer Genome Atlas (TCGA) database. MAGE-A11 expression was assessed in a well-characterized series of HNSCC (N = 75) with long-term follow-up and 10 cases of adjacent non-cancerous tissues, which were diagnosed between 2013 and 2014, by using immunohistochemistry. The correlation between MAGE-A11 expression and clinicopathological factors was analyzed. Kaplan–Meier and Cox regression analyses were used to assess the prognostic significance of MAGE-A11 expression among HNSCC patients.
Results: The results showed that MAGE-A11 mRNA expression was increased in HNSCC tissues compared to “normal” tissues (P < 10− 12). MAGE-A11 protein expression was not correlated with lymph node status, relapse, age, gender, histological grade, differentiation, clinical stage, tumor size, radiotherapy or chemotherapy. The patients with high MAGE-A11 expression had lower 5-year overall survival (OS) rates than those with low MAGE-A11 expression as determined using the Kaplan–Meier method. The univariate and multivariate analyses confirmed that elevated MAGE-A11 was an independent prognostic factor for the OS of HNSCC patients.
Conclusion: These findings indicate that MAGE-A11 may be a valuable diagnostic or prognostic marker as well as a potential molecular therapy target for HNSCC patients.
Keywords: Melanoma-associated antigen-A11, immunohistochemistry, head and neck squamous cell carcinoma, prognosis
Introduction
Head and neck squamous cell carcinomas (HNSCCs) rank as the sixth most common neoplasm in the world, about 600,000 new cases worldwide each year, with 40–50% mortality.1 More than 90% of the oral malignancies are squamous cell carcinomas, which arise from the epithelial lining of the oral cavity. Although cancer therapy has improved rapidly over the past decade, particularly advanced radiotherapy with or without chemotherapy, enhanced surgical procedures and immunotherapy, questions remain unanswered regarding accurate and effective biomarkers of disease.2,3 It is reported that Melanoma-associated antigen-A (MAGE-A) proteins are highly expressed in oral squamous cell carcinoma (OSCC) and are promising targets for cancer immunotherapy.4,5 Several authors have already described significantly lower 5-year survival rates of patients with certain types of cancers that express MAGE-A antigens, and there is also a study that reveals that MAGE-A3, -A4, -A5, -A9, -A11 are factors that are related to metastatic tendencies.6–8
Melanoma-associated antigen-A11 (MAGE-A11), as a member of the MAGE-A family, which belongs to Cancer/testis antigens (CTA), is an X-linked and primate-specific steroid hormone receptor transcriptional coregulator and proto-oncogenic protein expressed at low levels in normal human reproductive tract tissues and at higher levels in castration-resistant prostate cancer where it is required for androgen-dependent cell growth.9–11 Melanoma-associated antigen (MAGE) has been identified in a variety of types of cancer.12,13 The expression of several MAGE subgroups is correlated with poor prognosis and chemotherapeutic resistance.14 It was reported that MAGE-A11 is an independent poor prognostic marker for esophageal squamous cell carcinoma (ESCC).15 And frequently expressed in breast cancer which is associated with poor prognosis as well.16 MAGE-A11 is activated through TFCP2/ZEB1 binding sites demethylation as well as histone modification.17,18 Overexpression of MAGE-A11 changed a variety of gene expressions, which was associated with various cell functions such as protein ubiquitination, cell proliferation and apoptosis, tumor invasion and metastasis.
In this study, we aimed to reveal the expression levels and the prognostic relevance of MAGE-A11 in HNSCC tissues by immunohistochemistry in a random sample of 75 patients with HNSCC, exploring how it affects clinicopathological features and patient survival. Furthermore, we aimed to find a new molecular target that could be used as prognostic tools for improving the follow-up care of patients with HNSCC.
Materials and Methods
Parents and Ethics Statements
The formalin-fixed, paraffin-embedded specimens used for immunohistochemistry were collected from 75 HNSCC patients after surgical resections with no preoperative chemotherapy or radiotherapy in the Oral Hospital of China Medical University from 2013 to 2014. Data were collected from the patients’ operative and pathological reports, and follow-up data were retrieved from the clinical database. Clinicopathological data and patient characteristics were obtained from medical archives by retrospective analysis. The Human Research Ethical Committee of the China Medical University Affiliated Stomatological Hospital had approved that the use of these tissue samples for the study is reasonable. We had got the agreement of all the patients included in the study that their tumor samples could be used for the purpose of investigation during their initial diagnosis. Furthermore, written consents showing that we obtained the patients’ willingness to take part in this study from all participants. This study was conducted in accordance with the Declaration of Helsinki.
RNA Extraction and Real-Time PCR Assay
First, total RNA from 10 cases of HNSCC tissues and paired non-cancerous tissues were extracted by using TRIzol (Invitrogen, Carlsbad, CA, USA). Then, human immortal keratinocyte line (HaCaT), HNSCC cell lines (Cal27, Tca8113 and SCC9) were grown in a 6-well culture plate to 70–80% confluence before total RNA extraction with TRIzol (Invitrogen, Carlsbad, CA, USA) according to the manufacturer’s instructions. 500 ng total RNA were reverse transcribed to cDNA using QuantiTect Reverse Transcription Kit (TaKaRa, Shiga, Japan). The SYBR green dye (Takara, Shiga, Japan) was used for the amplification of cDNA. The mRNA levels of MAGE-A11, as well as that of the internal standard GAPDH, were measured by real-time quantitative PCR in triplicate using an Mx3000PTM Real-Time PCR System by Agilent (Stratagene, La Jolla, CA, USA). The specific primers used for these genes are listed in Table 1.
 |
Table 1 Sequence of Primers |
Immunohistochemical Staining
First, paraffin sections were taken from the specimens and cut into 4-μm thick sections. They were then added onto poly-lysine-coated slides and incubated at 65°C overnight. The incubated slides were then deparaffinized in xylene and rehydrated with graded alcohol. The next step was to retrieve the antigen using citrate buffer (pH 6.0) and store the slides in Tris buffered saline (TBS). In order to block endogenous peroxidase activity, 3% hydrogen peroxide was added to the slides. They were then incubated overnight at 4°C in rabbit polyclonal antibody (MAGE-A11) solution at 1:50 dilution. Finally, the slides were incubated with horseradish peroxidase-conjugated goat anti-rabbit immunoglobulin, and color was developed using the DAB Horseradish Peroxidase Color Development Kit (Maixin Co., Fuzhou, China).
Evaluation of Immunostaining
Two independent pathologists were separated to deal with the evaluation for positive DAB staining of all the immunoreactions. Exam every slide five times, and observed 100 cells during each examination using a medical microscope at 400 times magnification. There was positive immunostaining of the tumor cell membranes. Tissue samples stained for MAGE-A11 expression were classified into five categories and given a score from 0–5 according to the percentage of positively stained cells in each sample: “0” (0%), “1” (1–5%), “2” (5–25%), “3” (25–50%) and “4” (50–100%). Additionally, the staining intensity of tissue samples was used to divide them into four categories and assign them a score between 0–3: 0: negative, 1: weak, 2: moderate and 3: strong. Then, the sum of the first and second score was used to determine MAGE-A11 expression levels: 0–2, low expression and 3–7, high expression. In this way, HNSCC patients were divided into two groups: MAGE-A11-high and MAGE-A11-low patients.
Statistical Analysis
In this study, all statistical analyses were performed using the SPSS version 17.0 software (SPSS, Chicago, IL, USA). The possible connection between MAGE-A11 expression and clinicopathological features of HNSCC patients was examined using the χ2 test. Five-year Kaplan-Meier survival curves were generated, and the differences between the curves were estimated using the Log rank test. OS curves (overall survival), defined from HNSCC diagnosis to the date of death from any cause recorded were generated to determine the survival differences between the MAGE-A11-high and MAGE-A11-low patients. Based on the results of these two curves, the effects of MAGE-A11 expression on patient survival was examined using the hazards regression (HR), with the calculation of both univariate and multivariate Cox proportional HR models. It was considered statistically as significant differences when P values were less than 0.05.
Bioinformatics Analysis
We analyzed with different bioinformatics tools, including GEPIA (http://gepia.cancer-pku.cn)19 and UALCAN (http://ualcan.path.uab.edu).20 We used GEPIA, a web server for cancer and normal gene expression analyses and survival analyses, to extract MAGEA11 expression data from the Cancer Genome Atlas (TCGA) data portal and the Genotype-Tissue Expression (GTEx) database in 31 tumor types. All plotting features in GEPIA are developed using R (version 3.3.2) and Perl (version 5.22.1) programs. And a total of 519 HNSCC sample tissues and 44 “normal” samples taken adjacent to tumors were enrolled in this study. Regarding parameter settings, |log2FC| Cutoff of 1 and a q-value Cutoff of 0.01 were selected. UALCAN, a portal for facilitating tumor subgroup gene expression, can analysis MAGEA11 positively correlation genes in HNSCC.
Results
MAGE-A11 Is Overexpressed and Correlated with Poor Prognosis in HNSCC
We performed TCGA database GEPIA analysis to identify the expression level of MAGE-A11 mRNA in the most common cancers. As shown in Figure 1A, MAGE-A11 mRNA expression was especially upregulated in squamous cell carcinoma in head and neck squamous cell carcinoma (HNSCC) and lung squamous cell carcinoma (LUSC). The information of all patients was from TCGA HNSCC database and the patients included 519 cases of HNSCC patient tissues (Tumor) and 44 cases of non-tumor patient tissues (Normal) (***P < 0.001) (Figure 1B). Based on the expression of MAGE-A11 mRNA, we performed the Kaplan-Meier analysis to estimate patient’s DFS. As shown in Figure 1C, Kaplan-Meier survival curve showed that patients with high MAGE-A11 expression had worse DFS in HNSCC patients (P = 0.042). Bioinformatics analysis results showed that MAGE-A11 mRNA expression may positive relate with the expression of PRAME, DNAH14 and GTSF1 proteins by using UALCAN online tool (Figure 1D–F).
To further validate the mRNA expression of MAGE-A11, we assessed the expression levels of MAGE-A11 by qRT-PCR in 10 HNSCC tissues and paired non-cancerous tissues. As shown in Figure 2A the mRNA expression of MAGE-A11 had significant change (P < 0.0001). Nevertheless, the expression of MAGE-A11 in tumor cell lines (Cal27, Tca8113, SCC9) was increased compared with human immortal keratinocyte line (HaCaT) (Figure 2B). From the results, we found that the mRNA expression of MAGE-A11 was significantly upregulated in HNSCC tissues and cell lines.
High Expression of MAGE-A11 Is an Adverse Prognostic Factor in HNSCC Patients
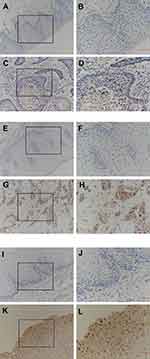
To validate the potential clinical utility of MAGE-A11 expression, a total of 85 paraffin-embedded samples were enrolled in this study, including 75 cases of HNSCC tissues and 10 cases of “Normal” tissues (the adjacent normal tissues/non-cancerous tissues). The detailed clinical characteristics include diagnosis at age, gender, tumor size, lymph node metastasis, TNM Stage, radiotherapy, chemotherapy, relapse, differentiation (Table 2). All patients were between the ages of 32 and 78 years (58.1 ± 8.47 years). With the paraffin-embedded HNSCC specimens staining, the expression of MAGE-A11 was high in 55 (73.3%) of the 75 cancer patients and low in 20 (26.7%) of the 75 patients, respectively. Further analysis revealed MAGE-A11 staining was predominantly in HNSCC tissue and the “Normal” (P < 0.001). As shown in Figure 3, MAGE-A11 was detected in cancer cells either in the nucleus, the nucleus & cytoplasm, and the cytoplasm, with the combined pattern of staining predominating. Table 3 summarizes the associations between MAGE-A11 expression and clinicopathological variables. There was no significant correlation between the MAGE-A11 expression and the clinicopathological parameters including age (P = 0.222), gender (P = 0.727), tumor size (P = 0.339), lymph node metastasis (P = 0.773), TNM stage (P = 0.861), radiotherapy (P = 0.624), chemotherapy (P = 0.569), relapse (P = 0.174), differentiation (P = 0.514).
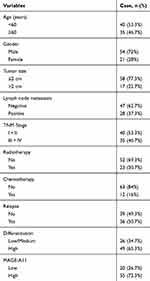 |
Table 2 Clinicopathological Data in HNSCC Patients |
 |
Table 3 Correlation of MAGE-A11 Immunoreactivity with Clinicopathological Variables in HNSCC Patients |
Then, we evaluated the prognostic power of MAGE-A11 protein on overall survival (OS) in 75 HNSCC patients. The patients with high MAGE-A11 expression had lower 5-year OS rates than those with low MAGE-A11 expression as determined using the Kaplan–Meier method (P = 0.027, Figure 4). To evaluate the impact of each variable on OS, univariate and multivariate Cox regression were used, as shown in Table 4. In the univariate analysis, the significant factors associated with OS included Tumor size (HR = 2.106, 95% CI = 1.042–4.26, P = 0.038), Lymph node metastasis (HR =2.082, 95% CI = 1.072–4.047, P = 0.03), TNM stage (HR =2.020, 95% CI = 1.016–4.019, P =0.045), Relapse (HR = 77.27, 95% CI = 10.475–570.006, P < 0.001) and MAGE-A11 expression (HR = 2.582, 95% CI =1.068–6.247, P = 0.035). In multivariate analyses, both MAGE-A11 expression (P = 0.002) and Relapse (P < 0.001) were associated with poor OS (Table 4).
 |
Table 4 Univariate and Multivariate Analysis of Overall Survival in HNSCC |
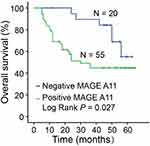 |
Figure 4 Kaplan–Meier survival curves for overall survival according to MAGE-A11 expression in HNSCC patients. P values were obtained by Log rank test (P = 0.027). |
Discussion
MAGE-A11, a member of the type I cancer-testis antigens, which expressed on the X chromosome, expressed at a higher level in a wide variety of human malignancies, including breast cancer, prostate cancer, head and neck cancer and esophageal squamous cell carcinoma.15,21,22 Importantly, previous studies showed that MAGE-A11, a key oncoprotein correlation with poor prognosis and survival in cancer, plays crucial role in the development of HNSCC.23,24 Hartmann et al shows that MAGE-A11 expression contributes to cisplatin resistance in head and neck cancer.25 We hypothesized that MAGE-A11 expression may be a predictor of malignant transformation in HNSCC.
Over the past decades, the MAGE protein family, which is a highly conserved group of proteins that share a common MAGE homology domain, has led to numerous insights that MAGEs were involved not only in stem cell differentiation and tumorigenesis progression, but also in invasion and metastasis.26,27 MAGE-A11, the unique steroid hormone receptor transcriptional coregulator among the type I MAGEs, is known to be involved in the transcriptional activity of androgen receptor (AR) by binding with p160 in prostate cancer.28–30 Furthermore, MAGE-A11, as a transcriptional activator of E2F1 and androgen receptor, interacts with retinoblastoma-related protein p107 and enhances prostate cancer cell growth.21,31,32 Mounting evidence indicated that MAGE-A11 was a key regulator of the survival of tumors by stabilizing HIF-1alpha levels. Recent studies have shown that MAGE-A11 can promote cell invasion and chemoresistance, though the targets underlying these processes are still poorly defined.
HNSCCs are among the most common cancers and high incidence and mortality worldwide, with being frequently diagnosed at advanced stage.33 The tobacco, alcohol, Epstein-Barr virus (EBV) and human papilloma virus (HPV) infection are widely perceived as the carcinogens, which could promote multiple sites carcinogenesis of the upper aerodigestive tract.34–38 The current treatment strategies of patients with HNSCC including surgery, radiation therapy (RT), chemotherapy with small molecule inhibitors or antibodies, immunotherapy, targeted therapy or combined modality treatments.39,40 Head and Neck cancers continue to pose a major treatment challenge for identification of biomarkers for the early detection and prognosis of HNSCC.
Accumulating evidence indicates that uncontrolled upregulation of MAGE-A11 protein is associated with abnormal tumor development and progression. Therefore, understanding the MAGE-A11 expression is important for cancer prevention and therapy. In this study, we used the data from the Cancer Genome Atlas dataset (TCGA-HNSCC) to analyze the expression and survival for MAGE-A11 using the online tool of GEPIA and UALCAN. The data analysis of TCGA-HNSCC showed that MAGE-A11 were highly expressed in HNSCC compared with the normal tissues and their expression level was correlated with the disease-free survival time. The expression of PRAME, DNAH14, GTSF1 genes are positively correlated with MAGE-A11 expression in HNSCC by UALCAN online tool. These indicated that MAGE-A11 may be a potential target for HNSCC. Furthermore, the gene expressions of MAGE-A11 were verified by quantitative real-time PCR in the patients and cell lines with HNSCC and we found that MAGE-A11 was up-expressed in HNSCC tissues and cell lines. Then, we incorporated staining intensity and percentage of immunopositivity tumor cells for the scoring of MAGE-A11 expression in HNSCC tissues. The results indicated that MAGE-A11 expression was significantly higher in cancerous tissues than adjacent-tumor tissues. Moreover, MAGE-A11 protein was found to be related to lymph node metastasis and recrudescence in the 75 HNSCC samples. The Cox regression analysis showed MAGE-A11 protein was detected as an independent prognostic factor for overall survival in HNSCC patients.
Taken together, we identified MAGE-A11 as a potential prognostic predictor for HNSCC patients. The study provides a set of relative target genes for future investigation into the molecular mechanisms and biomarkers. However, further function investigation is needed to explore the molecular mechanism of MAGE-A11 in HNSCC progression and metastasis.
Acknowledgments
This work was supported by the Project of National Natural Science Foundation of China (81902701), the Project of Natural Science Foundation of Liaoning Province (2015020528 and 20180530037), and College Students’ Innovation and Entrepreneurship Training Projects (201910159001 and 201910159084).
Disclosure
The authors declare that they have no competing interests.
References
1. Ferlay J, Soerjomataram I, Dikshit R, et al. Cancer incidence and mortality worldwide: sources, methods and major patterns in GLOBOCAN 2012. Int J Cancer. 2015;136(5):E359–E386. doi:10.1002/ijc.29210
2. Moskovitz J, Moy J, Ferris RL. Immunotherapy for head and neck squamous cell carcinoma. Curr Oncol Rep. 2018;20(2):22. doi:10.1007/s11912-018-0654-5
3. Graff-Dubois S, Faure O, Gross DA, et al. Generation of CTL recognizing an HLA-A*0201-restricted epitope shared by MAGE-A1, -A2, -A3, -A4, -A6, -A10, and -A12 tumor antigens: implication in a broad-spectrum tumor immunotherapy. J Immunol. 2002;169(1):575–580. doi:10.4049/jimmunol.169.1.575
4. Schooten E, Di Maggio A, van Bergen En Henegouwen PMP, Kijanka MM. MAGE-A antigens as targets for cancer immunotherapy. Cancer Treat Rev. 2018;67:54–62. doi:10.1016/j.ctrv.2018.04.009
5. Zajac P, Schultz-Thater E, Tornillo L, et al. MAGE-A antigens and cancer immunotherapy. Front Med. 2017;4:18. doi:10.3389/fmed.2017.00018
6. Sang M, Wu X, Fan X, Sang M, Zhou X, Zhou N. Multiple MAGE-A genes as surveillance marker for the detection of circulating tumor cells in patients with ovarian cancer. Biomarkers. 2014;19(1):34–42. doi:10.3109/1354750X.2013.865275
7. Suzuki S, Sasajima K, Sato Y, et al. MAGE-A protein and MAGE-A10 gene expressions in liver metastasis in patients with stomach cancer. Br J Cancer. 2008;99(2):350–356. doi:10.1038/sj.bjc.6604476
8. Miyashiro I, Kuo C, Huynh K, et al. Molecular strategy for detecting metastatic cancers with use of multiple tumor-specific MAGE-A genes. Clin Chem. 2001;47(3):505–512. doi:10.1093/clinchem/47.3.505
9. Wilson EM. Androgen receptor molecular biology and potential targets in prostate cancer. Ther Adv Urol. 2010;2(3):105–117. doi:10.1177/1756287210372380
10. Minges JT, Su S, Grossman G, et al. Melanoma antigen-A11 (MAGE-A11) enhances transcriptional activity by linking androgen receptor dimers. J Biol Chem. 2013;288(3):1939–1952. doi:10.1074/jbc.M112.428409
11. Wilson EM. Analysis of interdomain interactions of the androgen receptor. Methods Mol Biol. 2011;776:113–129.
12. Raghavendra A, Kalita-de Croft P, Vargas AC, et al. Expression of MAGE-A and NY-ESO-1 cancer/testis antigens is enriched in triple-negative invasive breast cancers. Histopathology. 2018;73(1):68–80. doi:10.1111/his.2018.73.issue-1
13. Veit JA, Heine D, Thierauf J, et al. Expression and clinical significance of MAGE and NY-ESO-1 cancer-testis antigens in adenoid cystic carcinoma of the head and neck. Head Neck. 2016;38(7):1008–1016. doi:10.1002/hed.v38.7
14. Xie C, Subhash VV, Datta A, et al. Melanoma associated antigen (MAGE)-A3 promotes cell proliferation and chemotherapeutic drug resistance in gastric cancer. Cell Oncol. 2016;39(2):175–186. doi:10.1007/s13402-015-0261-5
15. Sang M, Gu L, Liu F, et al. Prognostic significance of MAGE-A11 in esophageal squamous cell carcinoma and identification of related genes based on DNA microarray. Arch Med Res. 2016;47(3):151–161. doi:10.1016/j.arcmed.2016.06.001
16. Lian Y, Sang M, Ding C, et al. Expressions of MAGE-A10 and MAGE-A11 in breast cancers and their prognostic significance: a retrospective clinical study. J Cancer Res Clin Oncol. 2012;138(3):519–527. doi:10.1007/s00432-011-1122-x
17. Gu L, Sang M, Li J, et al. Expression and prognostic significance of MAGE-A11 and transcription factors (SP1,TFCP2 and ZEB1) in ESCC tissues. Pathol Res Pract. 2019;215(7):152446. doi:10.1016/j.prp.2019.152446
18. Liu S, Liu F, Huang W, et al. MAGE-A11 is activated through TFCP2/ZEB1 binding sites de-methylation as well as histone modification and facilitates ESCC tumor growth. Oncotarget. 2018;9(3):3365–3378. doi:10.18632/oncotarget.22973
19. Tang Z, Li C, Kang B, Gao G, Li C, Zhang Z. GEPIA: a web server for cancer and normal gene expression profiling and interactive analyses. Nucleic Acids Res. 2017;45(W1):W98–W102. doi:10.1093/nar/gkx247
20. Chandrashekar DS, Bashel B, Balasubramanya SAH, et al. UALCAN: a portal for facilitating tumor subgroup gene expression and survival analyses. Neoplasia. 2017;19(8):649–658.
21. Su S, Parris AB, Grossman G, Mohler JL, Wang Z, Wilson EM. Up-regulation of follistatin-like 1 by the androgen receptor and melanoma antigen-A11 in prostate cancer. Prostate. 2017;77(5):505–516. doi:10.1002/pros.v77.5
22. Hou SY, Sang MX, Geng CZ, et al. Expressions of MAGE-A9 and MAGE-A11 in breast cancer and their expression mechanism. Arch Med Res. 2014;45(1):44–51. doi:10.1016/j.arcmed.2013.10.005
23. Hartmann S, Zwick L, Maurus K, et al. Melanoma-associated antigen A11 reduces erlotinib and afatinib efficacy in head and neck cancer. J Craniomaxillofac Surg. 2018;46(3):492–497. doi:10.1016/j.jcms.2017.12.014
24. Karia BTR, Zamuner FT, Carlin V, de Oliveira CZ, Carvalho AL, Vettore AL. Expression and prognostic relevance of GAGE1 and XAGE1 cancer/testis antigens in head and neck squamous cell carcinoma. Curr Mol Med. 2017;17(10):707–717. doi:10.2174/1566524018666180322162145
25. Hartmann S, Zwick L, Scheurer MJJ, et al. MAGE-A11 expression contributes to cisplatin resistance in head and neck cancer. Clin Oral Investig. 2018;22(3):1477–1486. doi:10.1007/s00784-017-2242-8
26. Weon JL, Potts PR. The MAGE protein family and cancer. Curr Opin Cell Biol. 2015;37:1–8. doi:10.1016/j.ceb.2015.08.002
27. Gordeeva O, Gordeev A, Khaydukov S. Expression dynamics of Mage family genes during self-renewal and differentiation of mouse pluripotent stem and teratocarcinoma cells. Oncotarget. 2019;10(35):3248–3266. doi:10.18632/oncotarget.26933
28. Minges JT, Grossman G, Zhang P, Kafri T, Wilson EM. Post-translational down-regulation of melanoma antigen-A11 (MAGE-A11) by human p14-ARF tumor suppressor. J Biol Chem. 2015;290(41):25174–25187. doi:10.1074/jbc.M115.663641
29. Willett CS, Wilson EM. Evolution of Melanoma antigen-A11 (MAGEA11) during primate phylogeny. J Mol Evol. 2018;86(3–4):240–253. doi:10.1007/s00239-018-9838-8
30. Askew EB, Bai S, Parris AB, Minges JT, Wilson EM. Androgen receptor regulation by histone methyltransferase suppressor of variegation 3-9 homolog 2 and melanoma antigen-A11. Mol Cell Endocrinol. 2017;443:42–51. doi:10.1016/j.mce.2016.12.028
31. Su S, Minges JT, Grossman G, Blackwelder AJ, Mohler JL, Wilson EM. Proto-oncogene activity of melanoma antigen-A11 (MAGE-A11) regulates retinoblastoma-related p107 and E2F1 proteins. J Biol Chem. 2013;288(34):24809–24824. doi:10.1074/jbc.M113.468579
32. Attwooll C, Lazzerini Denchi E, Helin K. The E2F family: specific functions and overlapping interests. EMBO J. 2004;23(24):4709–4716. doi:10.1038/sj.emboj.7600481
33. Leemans CR, Snijders PJF, Brakenhoff RH. The molecular landscape of head and neck cancer. Nat Rev Cancer. 2018;18(5):269–282. doi:10.1038/nrc.2018.11
34. Ribeiro IP, de Melo JB, Carreira IM. Head and neck cancer: searching for genomic and epigenetic biomarkers in body fluids – the state of art. Mol Cytogenet. 2019;12:33. doi:10.1186/s13039-019-0447-z
35. Koneva LA, Zhang Y, Virani S, et al. HPV integration in HNSCC correlates with survival outcomes, immune response signatures, and candidate drivers. Mol Cancer Res. 2018;16(1):90–102. doi:10.1158/1541-7786.MCR-17-0153
36. Gourd E. Concurrent chemotherapy improves outcomes in HNSCC. Lancet Oncol. 2018;19(7):e343. doi:10.1016/S1470-2045(18)30452-2
37. Brakenhoff RH, Wagner S, Klussmann JP. Molecular patterns and biology of HPV-associated HNSCC. Recent Results Cancer Res. 2017;206:37–56. doi:10.1007/978-3-319-43580-0_3
38. Jiang R, Ekshyyan O, Moore-Medlin T, et al. Association between human papilloma virus/Epstein-Barr virus coinfection and oral carcinogenesis. J Oral Pathol Med. 2015;44(1):28–36. doi:10.1111/jop.2014.44.issue-1
39. Kaidar-Person O, Gil Z, Billan S. Precision medicine in head and neck cancer. Drug Resist Updat. 2018;40:13–16. doi:10.1016/j.drup.2018.09.001
40. Przybylski K, Majchrzak E, Weselik L, Golusinski W. Immunotherapy of head and neck squamous cell carcinoma (HNSCC). Immune checkpoint blockade. Otolaryngol Pol. 2018;72(6):10–16. doi:10.5604/01.3001.0012.4367
 © 2020 The Author(s). This work is published and licensed by Dove Medical Press Limited. The full terms of this license are available at https://www.dovepress.com/terms.php and incorporate the Creative Commons Attribution - Non Commercial (unported, v3.0) License.
By accessing the work you hereby accept the Terms. Non-commercial uses of the work are permitted without any further permission from Dove Medical Press Limited, provided the work is properly attributed. For permission for commercial use of this work, please see paragraphs 4.2 and 5 of our Terms.
© 2020 The Author(s). This work is published and licensed by Dove Medical Press Limited. The full terms of this license are available at https://www.dovepress.com/terms.php and incorporate the Creative Commons Attribution - Non Commercial (unported, v3.0) License.
By accessing the work you hereby accept the Terms. Non-commercial uses of the work are permitted without any further permission from Dove Medical Press Limited, provided the work is properly attributed. For permission for commercial use of this work, please see paragraphs 4.2 and 5 of our Terms.