Back to Journals » Breast Cancer: Targets and Therapy » Volume 11
LXR/RXR pathway signaling associated with triple-negative breast cancer in African American women
Authors Torres-Luquis O, Madden K, N'dri NMR, Berg R , Olopade OF , Ngwa W, Abuidris D, Mittal S , Lyn-Cook B, Mohammed SI
Received 31 August 2018
Accepted for publication 15 October 2018
Published 20 December 2018 Volume 2019:11 Pages 1—12
DOI https://doi.org/10.2147/BCTT.S185960
Checked for plagiarism Yes
Review by Single anonymous peer review
Peer reviewer comments 2
Editor who approved publication: Professor Pranela Rameshwar
Odalys Torres-Luquis,1,2 Krystal Madden,3 N’sanh MR N’dri,1 Richard Berg,4 Olufunmilayo F Olopade,5 Wilfred Ngwa,6,7 Dafalla Abuidris,8 Suresh Mittal,1,2 Beverly Lyn-Cook,9 Sulma I Mohammed1,2
1Department of Comparative Pathobiology, Purdue University, West Lafayette, IN, USA; 2Purdue University Center for Cancer Research, Purdue University, West Lafayette, IN, USA; 3Department of Learning Sciences, University of Illinois at Chicago, Chicago, IL, USA; 4Department of Surgery, Indiana University Health, Lafayette, IN, USA; 5Center for Clinical Cancer Genetics and Global Health, Department of Medicine, University of Chicago, Chicago, IL, USA; 6Department of Radiation Oncology, Brigham and Women’s Hospital, Boston, MA, USA; 7Department of Radiation Oncology, Harvard Medical School, Boston, MA, USA; 8Department of Oncology, National Cancer Institute, University of Gezira, Gezira, Sudan; 9Division of Biochemical Toxicology, National Center for Toxicological Research, Jefferson, AR, USA
Background: Triple-negative breast cancer (TNBC) is more prevalent in African and African American (AA) women compared to European American (EA) women. African and AA women diagnosed with TNBC experience high frequencies of metastases and less favorable outcomes. Emerging evidence indicates that this disparity may in fact be the result of the uniquely aggressive biology of African and AA disease.
Purpose: To understand the reasons for TNBC in AA aggressive biology, we designed the present study to examine the proteomic profiles of TNBC and luminal A (LA) breast cancer within and across patients’ racial demographic groups in order to identify proteins or molecular pathways altered in TNBC that offer some explanation for its aggressiveness and potential targets for treatment.
Materials and methods: Proteomic profiles of TNBC, LA tumors, and their adjacent normal tissues from AA and EA women were obtained using 2-dimensional gel electrophoresis and bioinformatics, and differentially expressed proteins were validated by Western blot and immunohistochemistry. Our data showed that a number of proteins have significantly altered in expression in LA tumors compared to TNBC, both within and across patients’ racial demographic groups. The differentially overexpressed proteins in TNBC (compared to LA) of AA samples were distinct from those in TNBC (compared to LA) of EA women samples. Among the signaling pathways altered in AA TNBC compared to EA TNBC are innate immune signaling, calpain protease, and pyrimidine de novo synthesis pathways. Furthermore, liver LXR/RXR signaling pathway was altered between LA and TNBC in AA women and may be due to the deficiency of the CYP7B1 enzyme responsible for cholesterol degradation.
Conclusion: These findings suggest that TNBC in AA women enriched in signaling pathways that are different from TNBC in EA women. Our study draws a link between LXR/RXR expression, cholesterol, obesity, and the TNBC in AA women.
Keywords: breast, triple-negative breast cancer, African American, European American, luminal breast cancer, luminal
Introduction
Triple-negative breast cancer (TNBC) is a breast cancer subtype that does not express estrogen receptors (ER) and progesterone receptors (PR) and lacks human EGF receptor-2 (HER-2) amplification.1 Although TNBC constitutes small percentage (10–20%) of all invasive breast cancers in women living in USA,2 it has very aggressive characteristics and distinct metastatic pattern and lacks targeted therapies.3 Epidemiological evidence showed that TNBC is more prevalent in young African and African American (AA) women compared to European American (EA) women and disproportionally lead to their death.4 Previous research attributed this disparity in death rates to a various socioeconomic factors including income, co-morbid disease, and limited access to health care and medical treatment.5 However, emerging evidence indicates that these disparities may in fact be due to the uniquely aggressive biology of the disease in African and AA. Results of studies comparing the biological differences between TNBC in AA and EA women were conflicted. Additional research has suggested that the interaction between the disparities and signaling pathways may promote TNBC’s aggressive biology and genomic instability.6,7 Pathways that included cytoskeletal remolding, cell adhesion, epithelial msenchymal transition, and Wnt/β-catenin were shown to be overrepresented in TNBC in AA and East African women. Activation of Wnt/β-catenin pathway was suggested as the pathway that may contribute to the more aggressive TNBC phenotype in women of African origin.8
Despite the knowledge gained from previous studies, these comparative investigations have not yet examined the gene or protein expression of TNBC and LA tumor within patient’s racial demographic to identify the differences that may have contributed to TNBC’s aggressiveness. Compared to TNBC, LA tumors represent the commonest breast cancer subtype as it forms about 50–60% of all breast cancer and is characterized by ER and PR expression and negative HER-2 amplification.4 LA tumors are characterized by lower level of proliferation-related genes as well as low histological grade, low degree of nuclear pleomorphism, and low mitotic activity.9 Unlike TNBC, patients with LA breast cancer have good prognoses, significantly lower relapse rates, and hormonal therapy treatment options.10
We designed the present study to examine the proteomic profiles of TNBC and LA breast cancer within patients’ racial demographic groups (AA women) and across patients’ racial demographics (AA vs EA). The goal is to identify proteins or molecular pathways altered in TNBC that offer explanation for its aggressiveness as well as potential targets for the treatment in African and AA women.
Materials and methods
Chemicals and reagents
The following reagents were purchased from Sigma-Aldrich Co. (St Louis, MO, USA): Coomassie Brilliant Blue R-250, dithiothreitol (DTT), urea, trypsin, glycerol, glacial acetic acid, alpha-cyano-4-hydroxycinnamic acid, acetonitrile, sodium carbonate, HAuCl4, casein, and Ponceau S. We have purchased ReadyStrip (IPG strip pH 4–7) from Bio-Rad Laboratories Inc. (Hercules, CA, USA). Primary mouse monoclonal antibody or rabbit polyclonal antibodies against gelsolin, calpain, peroxiredoxin-2 (Abcam, Cambridge, UK), PEBP, LDH-B, crystalline (Abgent, San Diego, CA, USA), anexxin-2 and LXRα and CRY7B1 (Novus, Littleton, CA, USA), and clusterin (R&D systems, Inc., Minneapolis, MN, USA) were used. We have purchased Mayer hematoxylin from Richard-Allan Scientific (Thermo Fisher Scientific, Waltham, MA, USA) and immunohistochemistry (IHC) reagents from Biocare Medical (Concord, CA, USA). All cell lines were purchased from the American Type Culture Collection (ATCC) (Manassas, VA, USA).
Breast cancer tissue preparation
Treatment-naive fresh or frozen invasive tumor and matched adjacent normal tissue samples (Table 1) were obtained from patients diagnosed with and undergone surgical removal of their invasive breast cancer at Indiana Health Hospital at Lafayette or from Indiana University Cancer Center Tissue Procurement and Distribution Core. Furthermore, additional breast cancer samples from AA were obtained from the University of Chicago. The Institutional Board Review Committee of Indiana Health and Purdue University and University of Chicago approved the use of these samples. All patients whose tissue samples were used in this research had provided written informed consent, and this was in accordance with the Declaration of Helsinki. The obtained samples were age-matched from self-identified AA and EA women for a total of 153 invasive cancer and normal samples. Pathological features and hormone and HER-2 amplification statuses were obtained from the pathology report. Tissue blocks/slides from invasive breast cancer that were formalin-fixed paraffin embedded (FFPE) from AA women and women from Sudan were obtained from Indiana University Cancer Center Tissue Procurement and Distribution Core (n=40; 20 each of LA and TNBC) and from National Cancer Institute, University of Gezira, Sudan (n=100), respectively, for biomarkers’ validation by IHC.
  | Table 1 Patients’ characteristics Abbreviations: LA, luminal A; TNBC, triple-negative breast cancer. |
Breast cancer tissues’ protein extraction
Approximately, 500 mg of each breast tissue was quickly thawed. To remove residual blood, the tissues were washed in ice-cold “salt-free” phosphate buffer (8 mM Na2HPO4, 2 mM KH2PO4). The samples were then homogenized in lysis buffer (“salt free” phosphate buffer pH 7.5, 0.5% Triton X-100, 1 mM phenylmethylsulfonyl fluoride (Sigma-Aldrich Co., St Louis, MO, USA), 15 µg/mL aprotinin, 10 µg/mL leupeptin, 10 µg/mL pepstatin, 100 µg/mL DNAse 1, 25 µg/mL RNAse A, 5 mM MgCl2) and centrifuged at 20,000¥ g for 15 minutes at 4°C. Amersham 2-D Quant Kit was used to determine the concentration of proteins. The proteins were precipitated using the trichloroacetic acid (Sigma-Aldrich Co., St Louis, MO, USA) /acetone precipitation method, and pellets were suspended in urea solution (9 M urea, 4% Igepal, 1% DTT, and 2% carrier ampholytes).
2-Dimensional gel electrophoresis
We performed 2-dimensional gel electrophoresis according to Li et al.11 Three gels per sample were prepared. Briefly, about 200 µg of protein of each sample was concentrated on isoelectric focusing tube gels (3.3% acrylamide, 9 M urea, 2% Igepal, 2% carrier ampholytes, pH 4–8) using the predetermined voltage program (500 V for 1 hour, 750 V for 1 hour, 1,000 V for 1 hour, and 1,400 V for 18.75 hours for a total of 28,500 V/hour). After that, tubes were loaded onto slab gels (linear gradient from 11 to 19%) in a Protean plus Dodeca Cell (Bio-Rad Laboratories Inc.) and the machine was switched to 160 V for 18 hours at 80°C. The gels were then fixed in a fixing solution (50% ethanol/2% phosphoric acid) overnight and then washed and stained by the Coomassie solution (methanol/17% ammonium sulfate/3% phosphoric acid and Coomassie Blue G-250). The gels were then washed and imaged using the GS-800 Calibrated Imaging Densitometer (Bio-Rad Laboratories Inc.). PDQuest software (Bio-Rad Laboratories Inc.) was used for image analysis. Individual protein abundances were determined by Student’s t-test using the PDQuest software.
Mass spectrophotometry analyses
For mass spectrophotometry analysis, significantly expressed protein determined by PDQuest analysis as described earlier was cut from the gel, destained with 50 mM ammonium bicarbonate first and followed by 50% acetonitrile and 100% acetonitrile, then, reduced with 10 mM DTT, alkylated with 55 mM iodoacetamide, and washed with 50 mM ammonium bicarbonate and 100% acetonitrile. The samples were digested with trypsin overnight at 37°C, and the peptides were then extracted and analyzed by matrix assisted laser desorption/ionisation - time of flight mass spectrometry (MALDI-TOF-MS) using a MicroMass M@LDI System (MicroMass) after being calibrated using peptide standards. ProteinLynx (MicroMass) was used to generate the mass list, which then submitted to Profound for database searches. A z score of 1.65 was obtained, which corresponds to the 95th percentile. The score was used as a threshold for positive identification of selected proteins.
IHC
IHC was performed according to Li et al.11 Approximately 5 µm of breast tissue sections were cut from FFPE tissues and mounted on positively charged SuperFrost slides (Thermo Fisher Scientific, Waltham, MA, USA). The tissue sections were then processed and stained with primary mouse monoclonal antibody or rabbit polyclonal antibodies PRDX2 (1:500), calpain (1:100), CYP7B1 (1:500), and LXRα (1:100) and visualized according to the manufacturer’s protocol (Biocare Medical). The antibodies used are CYp7B1. The grading scale of 0–3 (0, no staining; 1, equivocal staining; 2, moderate-to-intense staining; 3, highest intensity staining) was used to determine the intensity of each protein.
Western blot analysis
Western blot analysis was performed according to Mohammed et al.12 To extract protein from cell lines, lysis buffer was added to cultured cells at 75% confluence and protein was transferred to tubes and centrifuged. The protein concentration for each sample was determined using the Pierce bicinchoninic acid (BCA) Protein Assay Kit (Thermo Fisher Scientific). Proteins were separated by 10% SDS-PAGE and transferred onto nitrocellulose membrane. Detection was performed with primary anti-antibodies gelsolin, calpain, PRDX2, CRYAB, LDH-B, and PEBP2 at dilution at 1:1,000 and secondary antibody mouse or rabbit conjugated with horseradish peroxidase at 1:20,000 dilution. Bound complexes were then detected using the enhanced chemiluminescent system (Thermo Fisher Scientific). Equal loading was confirmed using β-actin (1:2,000) (Sigma-Aldrich Co.).
Ingenuity pathway analysis (IPA)
To determine the most relevant biological mechanisms, interaction networks, and functions of the differentially expressed proteins, proteins altered in expression between each category, hormonal and race status, or together list were submitted to Ingenuity Pathway Knowledge Base (Ingenuity System, Mountain View, CA, USA) and analyzed.
Statistical analysis
Statistical analysis was performed according to Li et al.11 Briefly, two-sample paired t-test was used to compare each group set using the log of the spot intensity. Zero was used to signify the lack of intensity if no spot was seen. We have used Bonferroni correction to adjust for multiple comparisons at the P-value of 0.05.
Results
The proteome landscape of LA tumors and TNBC in AA and EA women shows some similarity
The concentration of the protein extracted from AA and EA women TNBC and LA tissues and their normal counter parts was determined so as quantitatively analyze these samples using proteomic analysis. For all samples, we used only 200 mg of protein to balance the sensitivity concerns regarding gel staining and mass spectrometry analysis. During the analysis, we used the images to compare paired samples by hormonal and race status or both. Doing so, we were able to match (79%) all spots and found many to be differentially expressed with the Student’s t-test using the PDQuest software. We considered the protein to be significantly differentially overexpressed or down expressed at a fold change of 1.5 and a P-value of <0.05. Representative gels showing the proteome expression landscapes of breast cancer compared by hormone and race status are shown in Figure 1, and some selected differentially expressed spots between AA and EA women regardless of hormonal status are shown in Figure 2. In this study, to eliminate possible false positives and give a more stringent P-value, Bonferroni correction (P=0.005) was used. We used MALDI–TOF-MS to identify significantly differently expressed proteins between TNBC and LA in AA and EA women.
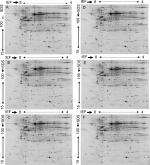  | Figure 1 Representative 2-DE gel images of protein profiles of invasive breast carcinoma. Notes: (A) TNBC vs LA in African American women, (B) African American vs European American women breast cancer samples, (C) TNBC in African American vs European American women, (D) TNBC vs LA in European American women, (E) TNBC vs LA regardless of race, and (F) LA in African American women vs LA in European American women. Proteins were separated by IEF as the first dimension, using 24 cm tube gels (pH 4–8), and linear gradient gel (11–19%) as the second dimension. The protein spots were cut from the gel, tryptically digested, and identified via MALDI–MS. Significantly expressed spots are posted in Table 2 along with their individual PDQuest spot number assignment and other data. Abbreviations: DE, 2-dimensional gel electrophoresis; LA, luminal A; MALDI-MS, matrix assisted laser desorption/ionisation - mass spectrometry; TNBC, triple-negative breast cancer. |
Differentially expressed proteins in LA vs TNBC in AA and EA women are different
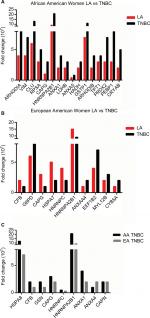
We found that 16 proteins were differentially expressed between TNBC and LA tissues from AA women (Figure 3A), while 11 proteins were differentially expressed between the two breast cancer subtypes in EA women (Figure 3B). However, only nine proteins were differentially expressed in TNBC in AA women compared to EA women. Vimentin, clusterin, HNRNP A2/B1, PRDX2, and crystalline were most overexpressed proteins between TNBC and LA in AA women. The protein differentially expressed between TNBC and LA tissues in AA women was different from that altered in expression between TNBC and LA tissues in EA women. Proteins that were differentially altered in TNBC and LA in EA women were HSP70, HNRNP C1/C2, HNRNP A2, and ELF-1B. However, HSP71 and HNRNP A2/B1 were the most altered proteins in expression when TNBC in AA women compared with that in EA women (Figure 3C). We show that TNBC from AA women characterized with high expression of vimentin while TNBC from EA women associated with high expression of myosin. These data suggested that TNBC in AA and EA women in our study belongs to the claudin-low subtype, however, expressing different proteins. These data also suggested that the landscape of TNBC in AA women is different from TNBC in EA women; however, not many proteins were altered in expression between AA and EA women TNBC.
Molecular pathway analysis of LA and TNBC proteins of AA women demonstrates upregulation of key nuclear receptors’ signaling and immunomodulation networks
We then submitted the identified overexpressed proteins gene identification to IPA to identify how these proteins are related to each other. We found that the top pathways altered between LA and TNBC in AA women included LXR/RXR, SUMO, IL-12, pyruvate fermentation to lactate, and RhoGDI signaling (Figure 4A). While the top pathways altered between TNBC and LA in EA women included acute phase response signaling, pentose phosphate pathway, complement system, and regulation of actin- based motility by Rho (Figure 4B). The top altered pathways in TNBC in EA and AA women are acute phase response signaling, integrin signaling, telomere extension, complement system, calpain protease, and pyrimidine ribonucleotide de novo biosynthesis (Figure 4C). These findings suggest that TNBC in AA women enriched in signaling pathways is different from TNBC in EA women.
Validation of selected differentially expressed proteins
We carried out a limited validation study in few of significantly differentially expressed proteins between TNBC and LA from AA tissue (Table 2) that included gelsolin, calpain, peroxiredoxin-2 (PRDX2), alpha-crystalline (CRYAB), lactate dehydrogenase β (LDH-B), and phosphatidylethanolamine binding protein-2 (PEBP2). For these validation studies, we have used both Western blot analysis and IHC. For Western blot analysis, we have used the following breast cancer cells: MDA468, a TNBC from a 51-year-old AA woman, HCC1500, a LA from a 31-year-old AA woman purchased from ATCC); and KTB 21 (normal). Gelsolin, calpain, PRDX2, and CRYAB were significantly (P-value <0.005) expressed in LA cells but not TNBC cells (Figure 5A and B). For IHC, we have used TNBC and LA tissues from AA women (n=40, 20 each) and African women (n=100). The IHC of PRDX2, calpain, CYP7B1, and LXRα was classified according to the score methods described in the “Materials and methods” section. PRXD2 had significantly (P<0.05) strong immunoreactivity in 85–95% of tumor cells in both LA and TNBC and were predominantly concentrated in the cytoplasm around the nucleus as shown in 75% of samples (Figure 5C and D). Calpain and LXRα showed high nonsignificant (P<0.05) expression in TNBC compared to LA tissues (Figure 5C and D). While CYP7B1 had strong immunoreactivity in tumor cells of LA and less expression in TNBC tissues (Figure 5C and D), CYP7B1 was significantly (P-value =0.005) expressed in LA relative TNBC cell lines (Figure 5E and F).
  | Table 2 Proteins altered in expression between TNBC and LA in African American and European American women Abbreviation: TNBC, triple-negative breast cancer. |
Discussion
In the era of personalized medicine, useful TNBC biomarkers and targeted therapeutic modalities do not exist. In this study, we have used proteomic analysis to identify proteins that account for the aggressive biology of TNBC in AA and African women. The precise knowledge of the proteome landscape of TNBC and LA in AA women compared to EA women may guide the development of new TNBC-targeted therapies. To our knowledge, the study described herein is the first to report on the differentially expressed proteins of TNBC and LA subtypes in AA and EA women using 2-dimensional gel electrophoresis coupled with protein identification via MALDI–TOF-MS and database analyses.
Our study defined the pattern of protein expression in TNBC and LA and their adjacent non-neoplastic tissues within and across racial demographics. We have identified a number of proteins that overexpressed in TNBC and LA in AA and EA patient samples. We did not identify a single protein that was significantly present in one subtype and missing in another; however, our work showed that certain proteins were increasingly upregulated in TNBC in AA patients than in EA patients. In addition, interestingly, the study showed that the differentially overexpressed proteins in the TNBC (compared to LA) of AA samples were distinct from the differentially expressed proteins in the TNBC (compared to LA) of EA samples. Our results agreed with recent transcriptomic analysis of data from white, black, and AA breast cancer patients’ normal and cancerous tissues from The Cancer Genome Atlas data repository showing that TNBC in white and black produces different abundances of mRNA, which are controlled in different ways and different regulators in the white and black or AA triple-negative patients.13 Therefore, we believe that these differences in transcriptome as well as proteome in our study may manifest as racial disparity in TNBC and could provide the rational for new diagnostics and targeted treatment for better overall survival rates in AA with TNBC.
A recently published study showed that the differentially expressed genes of age-matched TNBC in women of African descent and EA women were correlated with the Wnt–β-catenin pathway. This finding suggests that the activation of this pathway may contribute to the more aggressive TNBC phenotype of AA compared to the TNBC of EA women.6,7 However, our data showed increased representations of LXR/RXR, sumoylation, FXR/RXR, IL-12, and RhoGD1 signaling pathways in the LA (compared to TNBC) samples from AA women. Worth noting that, these LA and TBNC signaling pathways were not the same in the EA women samples.
LXR/RXR is a ligand-dependent transcriptional factor that is closely related to the nuclear receptors such as PPARs and FXR. The transcriptional activity of LXRs (two isoforms, LXRα and LXRβ) is dependent on the formation of heterodimers with retinoid X receptors (RXRs).14,15 The LXR/RXR plays an important physiological role in stimulating genes that regulate cholesterol, glucose, and fatty acid metabolism.16
Mice deficient in LXRα that were fed a high-cholesterol diet accumulated considerable amounts of cholesterol and lipid in their livers.17 Furthermore, mice deficient in LXRα receptors developed prostate hyperplasia lesions.18 LXRα deficiency caused prostate cancer cell line proliferation and survival in vitro and in vivo in animal models.19,20 In our study, however, we found that LXRα was expressed in both the LA and TNBC tissues of AA women. Accordingly, LXRα may not contribute to the biological differences between the two breast cancer subtypes.
To elucidate, further, the role of LXRα in the LA and TNBC in AA women, we examined the signaling pathway downstream of the receptor. We demonstrated that CYP7B1 enzyme was expressed in LA cell lines from both EA women and AA women but not in TNBC cells. Reduced CYP7B1 expression, which breaks down 27-hydroxycholesterol (27HC), resulted in its increased levels in the extrahepatic tissues. LXR activation as a result of 27HC accumulation was reported to promote breast cancer ER-positive cell line proliferation in vitro and in vivo.21 Mammary glands and uteri of young female mice, that is CYP7B1–/–, have the characteristics of tissues consistently exposed to estrogen and have showed advanced onset of puberty and early menarche, evidence of the premature fatigue of ovarian function in these mice.22 After adjusting for the effects of age, tumor size, nodal status, and perioperative therapy, multivariate Cox regression modeling demonstrated that low CYP7B1 expression was associated with poor breast cancer survival outcomes.23
Increased 27HC accumulation was shown in postmenopausal, hypercholesterolemic, and obese women. Both increased cholesterol levels and obesity are associated with an increased risk of developing breast cancer and poor prognosis24,25; both factors were also suggested as potential drivers of aggressive TNBC in AA women.26 The National Health and Nutrition Examination Survey (NHANES III) reported that more than half of AA women aged over 40 years were obese and more than 80% were overweight.27 Numerous studies link the use of statins, cholesterol-lowering drugs, and improved breast cancer outcomes. Statin use by women with inflammatory breast cancer significantly improved progression-free survival rates.28 Generally speaking, cancer patients who used statins were found to have a lower risk of dying from cancer compared to those cancer patients who are not on statins.29
Conclusion
The proteins identified in this proteomic study of TNBC and LA tissue samples from AA and EA women were different between AA and EA and associated with lipid metabolism, inflammation, immune system, and cell structure signaling pathways. Among such signaling pathways was LXR/RXR, which was altered in the TNBC samples of AA women. The accumulation of cholesterol, more specifically the cholesterol metabolite 27HC in TNBC tissues due to the deficient metabolizing enzyme CYP7B1, is a possible contributing factor to the aggressiveness of TNBC and may offer a targetable marker for its treatment.
Acknowledgments
The authors thank Dr H Harikrishna Nakshatri for providing the African American Normal cells, KTB 21, and Dr Frank Weizmann, Cellular and Integrative Physiology, Indiana University School of Medicine, for performing the MALDI–TOF analysis.
Author contributions
OT-L, KM, and NMRN conducted various experiments including 2D gel electrophoresis, Western blot, and immunohistochemistry. RB, OFO, DA, and BL-C provided the African American and European American women breast cancer tissues and cell lines. SIM contributed to the study design and result interpretation. All authors contributed to data analysis, drafting and revising the article, gave final approval of the version to be published, and agree to be accountable for all aspects of the work.
Disclosure
The authors report no conflicts of interest in this work.
References
Foulkes WD, Smith IE, Reis-Filho JS. Triple-negative breast cancer. N Engl J Med. 2010;363(20):1938–1948. | ||
O’Toole SA, Beith JM, Millar EK, et al. Therapeutic targets in triple negative breast cancer. J Clin Pathol. 2013;66(6):530–542. | ||
Anders C. Carey LA. Understanding and treating triple-negative breast cancer. Oncol. 2008;22(11):1233–1243. | ||
Carey LA, Perou CM, Livasy CA, et al. Race, breast cancer subtypes, and survival in the Carolina Breast Cancer Study. JAMA. 2006;295(21):2492. | ||
Vona-Davis L, Rose DP. The influence of socioeconomic disparities on breast cancer tumor biology and prognosis: a review. J Womens Health. 2009;18(6):883–893. | ||
Field LA, Love B, Deyarmin B, Hooke JA, Shriver CD, Ellsworth RE. Identification of differentially expressed genes in breast tumors from African American compared with Caucasian women. Cancer. 2012;118(5):1334–1344. | ||
Martin DN, Boersma BJ, Yi M, et al. Differences in the tumor microenvironment between African-American and European-American breast cancer patients. PLoS One. 2009;4(2):e4531. | ||
Dietze EC, Sistrunk C, Miranda-Carboni G, O’Regan R, Seewaldt VL. Triple-negative breast cancer in African-American women: disparities versus biology. Nat Rev Cancer. 2015;15(4):248–254. | ||
Yersal O, Barutca S. Biological subtypes of breast cancer: Prognostic and therapeutic implications. World J Clin Oncol. 2014;5(3):412–424. | ||
Guarneri V, Conte P. Metastatic breast cancer: therapeutic options according to molecular subtypes and prior adjuvant therapy. Oncologist. 2009;14(7):645–656. | ||
Li J, Abraham S, Cheng L, et al. Proteomic-based approach for biomarkers discovery in early detection of invasive urothelial carcinoma. Proteomics Clin Appl. 2008;2(1):78–89. | ||
Mohammed SI, Ren W, Flowers L, et al. Point-of-care test for cervical cancer in LMICs. Oncotarget. 2016;7(14):18787–18797. | ||
Telonis AG, Rigoutsos I. Race disparities in the contribution of miRNA isoforms and tRNA-derived fragments to Triple-negative breast cancer. Cancer Res. 2018;78(5):1140–1154. | ||
Hong C, Tontonoz P. Liver X receptors in lipid metabolism: opportunities for drug discovery. Nat Rev Drug Discov. 2014;13(6):433–444. | ||
Bovenga F, Sabbà C, Moschetta A. Uncoupling nuclear receptor LXR and cholesterol metabolism in cancer. Cell Metab. 2015;21(4):517–526. | ||
Laurencikiene J, Rydén M. Liver X receptors and fat cell metabolism. Int J Obes. 2012;36(12):1494–1502. | ||
Peet DJ, Turley SD, Ma W, et al. Cholesterol and bile acid metabolism are impaired in mice lacking the nuclear oxysterol receptor LXR alpha. Cell. 1998;93(5):693–704. | ||
Kim HJ, Andersson LC, Bouton D, Warner M, Gustafsson JA. Stromal growth and epithelial cell proliferation in ventral prostates of liver X receptor knockout mice. Proc Natl Acad Sci U S A. 2009;106(2):558–563. | ||
Zhuang L, Kim J, Adam RM, Solomon KR, Freeman MR. Cholesterol targeting alters lipid raft composition and cell survival in prostate cancer cells and xenografts. J Clin Invest. 2005;115(4):959–968. | ||
Pommier AJ, Dufour J, Alves G, et al. Liver x receptors protect from development of prostatic intra-epithelial neoplasia in mice. PLoS Genet. 2013;9(5):e1003483. | ||
Bruss V, Lu X, Thomssen R, Gerlich WH. Post-translational alterations in transmembrane topology of the hepatitis B virus large envelope protein. EMBO J. 1994;13(10):2273–2279. | ||
Warner M, Andersson S, Gustafsso J-A. Gene Expression Regulation: Steroid Hormone Effects. In: Squire LR, editor. Encyclopedia of Neuroscience. Amsterdam: Elsevier; 2009:619. | ||
Wu Q, Ishikawa T, Sirianni R, et al. 27-Hydroxycholesterol promotes cell-autonomous, ER-positive breast cancer growth. Cell Rep. 2013;5(3):637–645. | ||
Després JP, Moorjani S, Tremblay A, et al. Relation of high plasma triglyceride levels associated with obesity and regional adipose tissue distribution to plasma lipoprotein-lipid composition in premenopausal women. Clin Invest Med. 1989;12(6):374–380. | ||
George SM, Irwin ML, Smith AW, et al. Postdiagnosis diet quality, the combination of diet quality and recreational physical activity, and prognosis after early-stage breast cancer. Cancer Causes Control. 2011;22(4):589–598. | ||
Lee E, McKean-Cowdin R, Ma H, et al. Characteristics of triple-negative breast cancer in patients with a BRCA1 mutation: results from a population-based study of young women. J Clin Oncol. 2011;29(33):4373–4380. | ||
Ogden CL, Flegal KM, Carroll MD, Johnson CL. Prevalence and trends in overweight among US children and adolescents, 1999–2000. JAMA. 2002;288(14):1728–1732. | ||
Brewer TM, Masuda H, Liu DD, et al. Statin use in primary inflammatory breast cancer: a cohort study. Br J Cancer. 2013;109(2):318–324. | ||
Vedel-Krogh S, Nielsen SF, Nordestgaard BG. Statin Use Is Associated with Reduced Mortality in Patients with Interstitial Lung Disease. PLoS One. 2015;10(10):e0140571. |
 © 2018 The Author(s). This work is published and licensed by Dove Medical Press Limited. The full terms of this license are available at https://www.dovepress.com/terms.php and incorporate the Creative Commons Attribution - Non Commercial (unported, v3.0) License.
By accessing the work you hereby accept the Terms. Non-commercial uses of the work are permitted without any further permission from Dove Medical Press Limited, provided the work is properly attributed. For permission for commercial use of this work, please see paragraphs 4.2 and 5 of our Terms.
© 2018 The Author(s). This work is published and licensed by Dove Medical Press Limited. The full terms of this license are available at https://www.dovepress.com/terms.php and incorporate the Creative Commons Attribution - Non Commercial (unported, v3.0) License.
By accessing the work you hereby accept the Terms. Non-commercial uses of the work are permitted without any further permission from Dove Medical Press Limited, provided the work is properly attributed. For permission for commercial use of this work, please see paragraphs 4.2 and 5 of our Terms.