Back to Journals » OncoTargets and Therapy » Volume 12
LSD1 regulates Notch and PI3K/Akt/mTOR pathways through binding the promoter regions of Notch target genes in esophageal squamous cell carcinoma
Authors Hou G, Zhao Q, Zhang M, Wang P, Ye H, Wang Y, Ren Y, Zhang J, Lu Z
Received 1 March 2019
Accepted for publication 21 May 2019
Published 2 July 2019 Volume 2019:12 Pages 5215—5225
DOI https://doi.org/10.2147/OTT.S207238
Checked for plagiarism Yes
Review by Single anonymous peer review
Peer reviewer comments 2
Editor who approved publication: Dr Sanjeev K. Srivastava
Guiqin Hou,1 Qi Zhao,1 Mengying Zhang,1 Peng Wang,2 Hua Ye,2 Yang Wang,1 Yandan Ren,1 Jianying Zhang2,3 Zhaoming Lu1
1School of Pharmaceutical Sciences, Zhengzhou University, Zhengzhou, People’s Republic of China; 2College of Public Health, Zhengzhou University, Zhengzhou, People’s Republic of China; 3Henan Academy of Medical and Pharmaceutical Sciences, Zhengzhou University, Zhengzhou, People’s Republic of China
Background: The aberrant activation of Lysine-specific demethylase 1(LSD1), Notch and PI3K/Akt/mTOR signaling pathways were frequently happened in many cancers, including esophageal squamous cell carcinoma (ESCC). However, the regulatory relationship between LSD1 and Notch as well as PI3K/Akt/mTOR pathways is still unclear.
Purpose: This study aimed to explore the regulatory effects and mechanisms of LSD1 on Notch and PI3K/Akt/mTOR pathway in ESCC.
Results: Firstly, we demonstrated that LSD1 and proteins in Notch and PI3K/Akt/mTOR pathway were expressed in ESCC cells. Secondly, inhibition of LSD1 by tranylcypromine (TCP) or shRNA could decrease the expressions of related proteins in Notch and PI3K/Akt/mTOR signaling pathways in ESCC cells. Finally, we found that LSD1 could bind to the promoter regions of Notch3, Hes1 and CR2, and the combinations between them were reduced by TCP in ESCC.
Conclusion: Summarily, this study indicated that LSD1 might positively regulate Notch and PI3K/Akt/mTOR pathways through binding the promoter regions of related genes in Notch pathway in ESCC.
Keywords: LSD1, Notch, PI3K/Akt/mTOR, esophageal squamous cell carcinoma
Introduction
Esophageal carcinoma is one of the most common tumors worldwide. It is classified as esophageal squamous cell carcinoma (ESCC) or esophageal adenocarcinoma (EAC) depending on the clinicopathological features. EAC is now the most common cancer type in Western countries, while ESCC is the predominant type in China.1,2 Due to its high mortality rate and poor prognosis, the 5-year survival rate of ESCC patients in China is only about 20%; although thishas improved with the progress of treatment strategies in recent years, it is still very low.3 Therefore, further exploration of the molecular mechanisms of tumorigenesis in ESCC is warranted to develop novel targeted therapeutic strategies.
Epigenetic changes, such as abnormal DNA methylation and modification of histone, were reported to be associated with the development of various cancers.4–6 Lysine-specific demethylase 1 (LSD1, KDM1A), the first identified histone demethylase, could specifically catalyze the demethylation of monomethyl and dimethyl H3K4 (H3K4me1/2) and H3K9 (H3K9me1/2).7 The overexpression of LSD1 was correlated with tumorigenesis, and numerous LSD1 inhibitors have been developed so far.8 Tranylcypromine (TCP), which irreversibly inhibits LSD1 by forming a covalent adduct with the flavin adenine dinucleotide co-factor, has been shown to inhibit tumor growth in xenograft models of breast cancer and oral squamous cell carcinoma.9,10
The Notch pathway is involved in various cellular processes such as cell growth and differentiation.11 In mammals, the Notch family consists of four Notch receptors (Notch1–4) and five ligands (Delta-like 1, 3 and 4; Jagged 1 and 2.12 The binding of the ligand with the Notch receptor triggers proteolytic cleavage to release Notch intracellular domain (NICD), which is then translocated into the nucleus and forms a transcriptional activation complex with proteins in the Mastermind-like family and CSL, and thus transcriptional activates target genes of Notch such as hairy/enhancer of split 1 (Hes1), Deltex1 (DTX1), c-Myc, CR2 and nuclear factor-kappa B (NF-κB).13,14 It is reported that Notch pathway interacts with other signaling pathways such as PI3K/Akt/mTOR, Wnt and Ras/MAPK to regulate tumorigenesis in many types of cancers,15,16 and activated PI3K (phosphoinositide 3-kinase) regulates cell survival and proliferation by stimulating its downstream factors such as Akt and mTOR.17
Several studies have revealed potential regulatory effects of LSD1 on the Notch pathway;18–21 for example, LSD1 modulates the Notch/ASCL1 axis by binding to the Notch1 locus in small-cell lung cancer.21 Some studies also reported that LSD1 could affect the mTOR pathway through regulating autophagy.22,23 However, the regulating effects of LSD1 on the Notch and mTOR pathways remain poorly understood in ESCC, which was just the problem we explored in this study.
Materials and methods
Reagents and antibodies
TCP was purchased from MedChemExpress (USA). Primary antibodies recognizing LSD1 (ab17721) and Hes1 (ab71559) were obtained from Abcam (UK), DTX1 (GTX112367) were obtained from GeneTex (USA), and H3 (9728S), Notch 1 (3608S), Notch 3 (3889S), H3K4me2 (3889S), PI3K (4228S), Rictor (9476S), p-Akt (Ser473) (4060S), Akt (2920S), p-mTOR (Ser2448) (5536S), p-mTOR (Ser2481) (2974S), mTOR (2983S), Raptor (2280S), p-p70S6K (Thr389) (9206S) and GAPDH (5174S) as well as the secondary antibodies were obtained from Cell Signaling Technology (USA). LSD1 shRNA and control shRNA vector were obtained from GenePharma Company (Shanghai, China).
Cell lines and cell culture
KYSE450, KYSE790, ECa109, EC9706 and TE-1 cells were obtained from Cell Bank of Type Culture Collection of the Chinese Academy of Sciences (Shanghai, China). As described before,24 cells were cultured in RPMI-1640 medium (Biological Industries, Israel) containing 10% fetal bovine serum (FBS) (Biological Industries, Israel) with a CO2 incubator at the condition of 5% CO2 and 37 °C.
Western blot
Total proteins and histone proteins of KYSE450, KYSE790, ECa109, EC9706 and TE-1 cells treated with 10 or 50 µM TCP at for 48 h were extracted, respectively, using total protein or histone protein extraction kit (Epigentek, USA). 30 µg of total proteins or 2 µg of histone proteins were separated on 8–10% SDS-PAGE gels and then electro-transferred to supported nitrocellulose membranes using a wet transferor. After the membranes were blocked with 5% skimmed milk for 2 h, then incubated with primary antibodies recognizing LSD1 (1:1000), Hes1 (1:1000), DTX1 (1:1000), H3 (1:1000), Notch1 (1:1000), Notch3 (1:1000), H3K4me2 (1:1000), PI3K (1:500), Rictor (1:1000), p-Akt (Ser473) (1:1000), Akt (1:1000), p-mTOR (Ser2448) (1:1000), p-mTOR (Ser2481) (1:1000), mTOR (1:1000), Raptor (1:500), p-p70S6K (Thr389) (1:500) and GAPDH (1:1000) diluted in 5% skimmed milk in PBST at 4 °C overnight, followed by incubation with the HRP-linked secondary antibodies at room temperature (RT) for 2 h. The membranes were rinsed three times with TBST between the incubations described above. Finally, the bands of specific proteins on the membranes were visualized with enhanced chemiluminescence (ECL) reagent (Thermo Scientific, USA) according to the manufacturer’s instructions. All measures of protein expression were repeated at least three times independently and the blots were analyzed using Image J software (NIH, USA).
Cell transfection and screening
ECa109 and EC9706 cells seeded in six-well plates were transfected with LSD1 shRNA or control shRNA vector, respectively, using Lipofectamine® 3000 reagent (Invitrogen, USA) according to the manufacturer’s protocols and cells stably expressing LSD1 shRNA or control shRNA were screened as described earlier.25 In brief, after ECa109 and EC9706 cells (2×105 cells per well) were cultured in a six-well plate for 24 h, the culture medium was changed as fresh serum-free medium (2 mL per well). Meanwhile, 5 µg LSD1 shRNA or control shRNA vector, respectively, was mixed with a mixture of 125 µL serum-free medium and 5 µL Lipofectamine™ 3000 reagent for 5 min at RT. Cells continued to be cultured for 8 h after the mixture was added into the wells, then the medium was replaced with RPMI-1640 medium containing 10% FBS for another 24 h. Subsequently, 2.5 μg/mL of puromycin was added into the wells for screening cells with LSD1 shRNA or control shRNA vector, and the untreated cells were as control. When all the control cells were dead, the screened cells were cultured with medium containing 2.5 μg/mL puromycin for another 2 d, and continued to be cultured and passaged using medium with 1.25 μg/mL of puromycin, the total protein of which was extracted for analyzing the expression of LSD1 and proteins in the Notch and PI3K/Akt/mTOR pathways by Western blot.
Chromatin immunoprecipitation assay
Chromatin immunoprecipitation (ChIP) assay was performed using chromatin immunoprecipitation kit (Millipore, Germany) according to the manufacturer’s instructions. In brief, ECa109 cells treated with 50 µM of TCP were fixed in 1% formaldehyde at RT for 10 min, and 125 mM of glycine was added for another 5 min. Subsequently, the cells were washed with cold PBS containing protease inhibitors and resuspended in lysate buffer containing 1% SDS and protease inhibitors. After the cell lysates were placed on ice for 8 min and centrifuged at 12,000 rpm for 10 min, the protein G-agarose magnetic beads were added into the supernatant and incubated at 4 °C for 1 h, then centrifuged at 1000 rpm for 1 min. One percent of the supernatant was retained at 4 °C as input solution, and the remaining supernatant for ChIP assay was incubated at 4 °C overnight with antibodies against RNA polymerase II, normal mouse IgG and LSD1, respectively, followed by the introduction of 60 µL protein G-agarose and incubation at 4 °C for 1 h. After being centrifuged at 4 °C, 1000 rpm for 1 min, the sediment obtained was washed successively with low-salt, high-salt, LiCl and TE buffer solutions, being incubated for 5 min and centrifuged 1000 rpm for 1 min during each washing to obtain the DNA-protein complex. Subsequently, 200 µL of elution buffer consisting of 10 µL of 20% SDS, 20 µL of 1 M NaHCO3 and 170 µL of sterile distilled water was added into the DNA-protein complex at RT for 30 min. At the same time, the same volume of elution buffer was added to the input solution and kept at 4 °C. Then, the DNA-protein complex with elution buffer was incubated with 8 µL of 5 M NaCl at 65 °C for 4 h and1 µL of RNase A at 37 °C for 30 min, followed by the addition of 1 µL of proteinase K, 4 µL of 0.5 M EDTA and 8 µL of 1 M Tris-Hcl and incubation at 45 °C for 1 h. Finally, phenol and ethanol were used for extracting DNA following the template of quantitative real-time PCR (qRT-PCR) described below.
qRT-PCR analysis
Quantity binding with LSD1 of Notch target gene promoters was detected by qRT-PCR analysis using the SYRB Green PCR Kit (Roche, USA). The DNA above was subjected to qRT-PCR analysis in 20 μL system with 10 μL SYBR Green Master Mix, 2 μL DNA template, 1 μL forward primer, 1 μL reverse primer and 6 μL double distilled water. Primers for different regions of Notch target gene promoter used in the present study were synthesized by Sangon Biotech (Shanghai, China) as shown in Table 1. The PCR condition was as follows: 95 °C for 10 min; followed by 45 cycles at 95 °C for 10 s, 60 °C for 10 s and 72 °C for 10 s; finally 95 °C for 10 s, 65 °C for 60 s and 97 °C for 1 s. Relative quantity of mRNA was calculated using the comparative cycle threshold method (2−ΔΔCt) as previously described.26,27 The Ct value of each ChIP DNA sample (ΔCt[correction ChIP]) was adjusted according to the formula: ΔCt[correction ChIP]=(Ct[ChIP]-(Ct[Input]-Log2 (Input dilution fraction))), and the %input value for each ChIP DNA sample was calculated according to the formula: 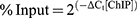 .
.
 |
Table 1 Primer sequences of Notch target genes promoter evaluated by qRT-PCR analysis |
Statistical analysis
All experiments in this study were performed at least three times and the results were analyzed by one-way analysis of variance (ANOVA) or Student’s t-test using SPSS 19.0 software. The results were presented as the mean ± standard deviation, with P<0.05 considered statistically significant.
Results
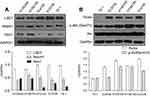
Expression of LSD1, proteins involved in Notch and mTOR pathway in ESCC cells
The expression levels of LSD1, related proteins in Notch and PI3K/Akt/mTOR signaling pathways in KYSE450, KYSE790, ECa109, EC9706 and TE-1 cell lines were detected by Western blot. As shown in Figure 1A, LSD1 was expressed in all the five ESCC cell lines, especially in ECa109 and EC9706 cell lines. The expressions of related proteins in the Notch pathway, such as Notch1 and Hes1, as well as in the mTOR pathway, such as Rictor and p-Akt (Ser473), were also observed in the five ESCC cell lines (Figure 1B), while the regulation relationship between LSD1 and the Notch and mTOR pathways warrants further exploration.
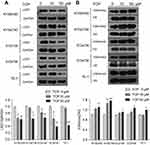
TCP inhibited the expression and function of LSD1 in ESCC cells
We determined firstly the effects of LSD1 inhibitor TCP on expression and function of LSD1 in KYSE450, KYSE790, ECa109, EC9706 and TE-1 cells by Western blot through treating cells with different concentrations of TCP (0, 10 and 50 μM) for 48 h. As shown in Figure 2A and B, after cells were treated with increasing doses of TCP, the expression of LSD1 clearly decreased while H3K4me2 increased. Compared to their control cells the decreasing ratios of LSD1 were 38.8%, 36.9%, 34.3%, 60.9% and 56.8%, respectively, and the increased ratios of H3K4me2 were 31.0%, 57.2%, 11.9%, 9.0% and 10.7%, respectively, in KYSE450, KYSE790, ECa109, EC9706 and TE-1 cells treated with 50 μM of TCP (P<0.05). These results suggest that TCP can not only inhibit the expression of LSD1, but also suppress the demethylating function of LSD1 in ESCC cells.
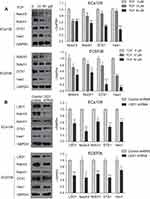
Downregulation of LSD1 inhibited Notch signaling pathway in ESCC cells
ECa109 and EC9706 cells, which had higher expression of LSD1 than other ESCC cell lines, were chosen to study the regulatory effects of LSD1 on the Notch signaling pathway in this study. After ECa109 and EC9706 cells were treated with different concentrations of TCP (0, 10 and 50 μM) for 48 h, the expressions of related proteins in the Notch signaling pathway were measured by Western blot. As shown in Figure 3A, TCP decreased the expressions of Notch3, Notch1, DTX1 and Hes1 in a dose-dependent manner in both cells (P<0.05). Compared with untreated cells, the decreasing ratios of Notch3, Notch1, DTX1 and Hes1 were 36.9%, 43.3%, 52.6% and 62.5%, respectively, in ECa109 cells, and 50.4%, 43.1%, 79.3% and 88.4%, respectively, in EC9706 cells at 50 μM of TCP, indicating that TCP could effectively inhibit the Notch signaling pathway.
In addition, to further determine the regulation effects of LSD1 on the Notch signaling pathway, after the expression of LSD1 was downregulated by LSD1 shRNA, the expressions of related proteins in the Notch signaling pathway were investigated by Western blot. As shown in Figure 3B, compared with the control shRNA group, the expression of LSD1 in the LSD1 shRNA group clearly decreased (P<0.05); and the decrease ratios were 43.8% and 46.4% in EC9706 and ECa109 cells, respectively, indicating that LSD1 shRNA was transfected successfully into cells and expressed stably in cells. After the expression of LSD1 in ESCC cells was knocked down, the expressions of Notch3, Notch1, DTX1 and Hes1 also decreased at ratios of 68.2%, 38.2%, 54.9% and 39.6%, respectively in ECa109 cells, and 53.7%, 41.8%, 53.3% and 30.2%, respectively in EC9706 cells (P<0.05). These findings demonstrate that inhibition of LSD1 by either TCP or shRNA decreased the expressions of related proteins in the Notch signaling pathway, suggesting that LSD1 might promote activation of the Notch signaling pathway in ESCC cells.
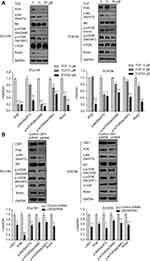
Downregulation of LSD1 inhibited the PI3K/Akt/mTOR signaling pathway in ESCC cells
We also investigated protein expression in the PI3K/Akt/mTOR signaling pathway by Western blot after ECa109 and EC9706 cells were treated with different concentrations of TCP (0, 10 and 50 μM) for 48 h. As shown in Figure 4A, along with the concentration increase of TCP, the expressions of PI3K, p-Akt (Ser473), p-mTOR (Ser2448), p-mTOR (Ser2481) and Rictor were decreased at ratios of 80.2%, 67.1%, 85.8%, 47.6%, 81.5% in ECa109 cells and 78.1%, 53.9%, 43.6%, 56.9%, 71.1% in EC9706 cells at 50 μM of TCP (P<0.05) compared with untreated cells, indicating that TCP could inhibit the PI3K/Akt/mTOR signaling pathway.
Moreover, downregulation of LSD1 by shRNA had a similar effect with TCP: the expressions of PI3K, p-Akt (Ser473), p-mTOR (Ser2448), p-mTOR (Ser2481) and Rictor decreased in the LSD1 shRNA group (P<0.05) compared with the control shRNA group, atratios of 80.5%, 41.4%, 30.4%, 28.9% and 44.7% in ECa109 cells and 55.9%, 54.1%, 24.1%, 33.7% and 41.3% in EC9706 cells, respectively (Figure 4B; P<0.05). These show that inhibition of LSD1 by either TCP or shRNA could inhibit the expressions of related proteins in the PI3K/Akt/mTOR signaling pathway, suggesting that LSD1 might contribute to activation of the PI3K/Akt/mTOR signaling pathway.
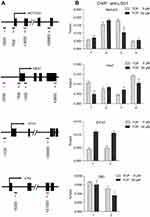
LSD1 binding the promoter regions of Notch target genes in ECa109 cells
The regulatory mechanisms of LSD1 to Notch signaling pathway were explored by ChIP analysis. As shown in Figure 5, after ECa109 cells were treated with 50 µM TCP for 48 h, the combined ability of LSD1 with the promoters of Notch3, HES1 and CR2 obviously decreased (P<0.05), but not DTX1. The combining site was at downstream 3000 bp of transcription start site (TSS) for Notch3, upstream 2200 bp and the downstream 2600 bp of TSS for Hes1, and downstream 21000 bp of TSS for CR2. These results indicated that LSD1 might regulate the Notch signaling pathway through binding the promoter regions of Notch target genes in ESCC cells.
Discussion
The unbalance of histone methylation and demethylation has been shown to be associated with the tumorigenesis of many cancers.6 Overexpression of LSD1, the first discovered histone demethylase, was involved in human carcinogenesis by regulating chromatin.8 The TCP-based LSD1 inhibitors have shown significant therapeutic effects in cancers and some of these have been tested in clinical trials, such as ORY1001 and GSK2879552, which indicated the potential of LSD1 as a therapeutic target for cancers.28 Recent studies have also reported that LSD1 is overexpressed in ESCC specimens, suggesting that it is a potential therapeutic target for ESCC.5,29 In this study, we also demonstrated clear expression of LSD1 in ESCC cell lines, and found that its expression and function could be inhibited by TCP.
Studies have shown that the Notch pathway is aberrantly activated in several human cancers, including ESCC,30 and LSD1 could negatively regulate the Notch signaling pathway by interacting with CoREST in the developing cerebral cortex or SIRT1 in neurogenesis;18,19 but LSD1 was also reported to be an integral component of the Notch-activating complex in the nucleus.20 Further, some reports proved that LSD1 could regulate the PI3K/Akt/mTOR pathway by combining the promoter region of tumor suppressor PTEN31 and inducing autophagy in ovarian cancer cells.23 In this study, we showed that inhibition of LSD1 decreased the expressions of related protein, not only in the Notch pathway (such as Notch3, Notch1, DTX1 and Hes1), but also in the PI3K/Akt/mTOR pathway such as PI3K, p-Akt (Ser473), p-mTOR (Ser2448), p-mTOR (Ser2481) and Rictor in ESCC cells, which indicated that LSD1 might have positively regulating effects on the Notch and PI3K/Akt/mTOR signaling pathways in ESCC. However, the molecular mechanism for this remains to be elucidated.
To explore the molecular mechanism of the regulation effects of LSD1 on the Notch and PI3K/Akt/mTOR pathways, we detected the combination of LSD1 with the promoter regions of Notch pathway targets Notch3, Hes1, DTX1 and CR2 by ChIP assay. The results clearly demonstrated combinations of LSD1 with the promoter regions of Notch3, Hes1 and CR2; and TCP prevented the combinations of LSD1 with them, indicating that LSD1 might positively regulate the Notch pathway through interacting with the promoter of Notch pathway target genes in ESCC. Many studies have demonstrated that the Notch pathway might act as upstream signaling to positively regulate the PI3K/Akt/mTOR signaling pathway, and inhibition of the Notch pathway by a γ-secretase inhibitor (GSI) decreased the activation of both Notch and Akt/mTOR pathways;32–35 thus we speculated that the molecular mechanism of LSD1 might positively regulate the PI3K/Akt/mTOR pathway through regulating the Notch pathway. Further investigation is needed.
Conclusion
Taken together, this study found early evidence that LSD1 might positively regulate the Notch and PI3K/Akt/mTOR signaling pathways in ESCC cells through combining to the promoter regions of Notch target genes. These findings might provide new insight into the effects of LSD1 and the Notch and mTOR pathways in the tumorigenesis of ESCC.
Acknowledgments
This study is supported by the Natural Science Foundation of Henan Province, China (No. 182300410312) and the National Science and Technology Major projects of China (No:2018ZX10302205).
Disclosure
The authors report no conflicts of interest in this work.
References
1. Murphy G, McCormack V, Abedi-Ardekani B, et al. International cancer seminars: a focus on esophageal squamous cell carcinoma. Ann Oncol. 2017;28(9):2086–2093. doi:10.1093/annonc/mdx279
2. Rubenstein JH, Shaheen NJ. Epidemiology, diagnosis, and management of esophageal adenocarcinoma. Gastroenterology. 2015;149(2):302–17 e1. doi:10.1053/j.gastro.2015.04.053
3. Cancer Genome Atlas Research N, Analysis Working Group: Asan U, Agency BCC, et al. Integrated genomic characterization of oesophageal carcinoma. Nature. 2017;541(7636):169–175. doi:10.1038/nature20805
4. Dawson MA, Kouzarides T. Cancer epigenetics: from mechanism to therapy. Cell. 2012;150(1):12–27. doi:10.1016/j.cell.2012.06.013
5. Hoshino I, Akutsu Y, Murakami K, et al. Histone demethylase LSD1 inhibitors prevent cell growth by regulating gene expression in esophageal squamous cell carcinoma cells. Ann Surg Oncol. 2016;23(1):312–320. doi:10.1245/s10434-015-4488-1
6. Di Stefano L, Walker JA, Burgio G, et al. Functional antagonism between histone H3K4 demethylases in vivo. Genes Dev. 2011;25(1):17–28. doi:10.1101/gad.1983711
7. Shi Y, Lan F, Matson C, et al. Histone demethylation mediated by the nuclear amine oxidase homolog LSD1. Cell. 2004;119(7):941–953. doi:10.1016/j.cell.2004.12.012
8. Hayami S, Kelly JD, Cho HS, et al. Overexpression of LSD1 contributes to human carcinogenesis through chromatin regulation in various cancers. Int J Cancer. 2011;128(3):574–586. doi:10.1002/ijc.25349
9. Ferrari-Amorotti G, Chiodoni C, Shen F, et al. Suppression of invasion and metastasis of triple-negative breast cancer lines by pharmacological or genetic inhibition of slug activity. Neoplasia. 2014;16(12):1047–1058. doi:10.1016/j.neo.2014.10.006
10. Wang Y, Zhu Y, Wang Q, et al. The histone demethylase LSD1 is a novel oncogene and therapeutic target in oral cancer. Cancer Lett. 2016;374(1):12–21. doi:10.1016/j.canlet.2016.02.004
11. Capaccione KM, Pine SR. The Notch signaling pathway as a mediator of tumor survival. Carcinogenesis. 2013;34(7):1420–1430. doi:10.1093/carcin/bgt127
12. Vanorny DA, Mayo KE. The role of Notch signaling in the mammalian ovary. Reproduction. 2017;153(6):R187–R204. doi:10.1530/REP-16-0689
13. Guo S, Liu M, Gonzalez-Perez RR. Role of Notch and its oncogenic signaling crosstalk in breast cancer. Biochim Biophys Acta. 2011;1815(2):197–213. doi:10.1016/j.bbcan.2010.12.002
14. Weng AP, Millholland JM, Yashiro-Ohtani Y, et al. c-Myc is an important direct target of Notch1 in T-cell acute lymphoblastic leukemia/lymphoma. Genes Dev. 2006;20(15):2096–2109. doi:10.1101/gad.1450406
15. Mittal S, Subramanyam D, Dey D, Kumar RV, Rangarajan A. Cooperation of Notch and Ras/MAPK signaling pathways in human breast carcinogenesis. Mol Cancer. 2009;23(8):128. doi:10.1186/1476-4598-8-128
16. Moghbeli M, Abbaszadegan MR, Golmakani E, Forghanifard MM. Correlation of Wnt and NOTCH pathways in esophageal squamous cell carcinoma. J Cell Commun Signal. 2016;10(2):129–135. doi:10.1007/s12079-016-0320-3
17. Hassan B, Akcakanat A, Holder AM, Meric-Bernstam F. Targeting the PI3-kinase/Akt/mTOR signaling pathway. Surg Oncol Clin N Am. 2013;22(4):641–664. doi:10.1016/j.soc.2013.06.008
18. Lopez CI, Saud KE, Aguilar R, et al. The chromatin modifying complex CoREST/LSD1 negatively regulates notch pathway during cerebral cortex development. Dev Neurobiol. 2016;76(12):1360–1373. doi:10.1002/dneu.22397
19. Mulligan P, Yang F, Di Stefano L, et al. A SIRT1-LSD1 corepressor complex regulates Notch target gene expression and development. Mol Cell. 2011;42(5):689–699. doi:10.1016/j.molcel.2011.04.020
20. Yatim A, Benne C, Sobhian B, et al. NOTCH1 nuclear interactome reveals key regulators of its transcriptional activity and oncogenic function. Mol Cell. 2012;48(3):445–458. doi:10.1016/j.molcel.2012.08.022
21. Augert A, Eastwood E, Ibrahim AH, et al. Targeting NOTCH activation in small cell lung cancer through LSD1 inhibition. Sci Signal. 2019;12:567. doi:10.1126/scisignal.aat8595
22. Li Y, Tao L, Zuo Z, et al. ZY0511, a novel, potent and selective LSD1 inhibitor, exhibits anticancer activity against solid tumors via the DDIT4/mTOR pathway. Cancer Lett. 2019;454:179–190. doi:10.1016/j.canlet.2019.03.052
23. Feng S, Jin Y, Cui M, Zheng J. Lysine-specific demethylase 1 (LSD1) inhibitor S2101 induces autophagy via the AKT/mTOR pathway in SKOV3 ovarian cancer cells. Med Sci Monitor. 2016;22:4742–4748.
24. Peng KZ, Ke Y, Zhao Q, et al. OP16, a novel ent-kaurene diterpenoid, potentiates the antitumor effect of rapamycin by inhibiting rapamycin-induced feedback activation of Akt signaling in esophageal squamous cell carcinoma. Biochem Pharmacol. 2017;140:16–27. doi:10.1016/j.bcp.2017.05.013
25. Hou G, Zhao Q, Zhang M, et al. Down-regulation of Rictor enhances cell sensitivity to PI3K inhibitor LY294002 by blocking mTORC2-medicated phosphorylation of Akt/PRAS40 in esophageal squamous cell carcinoma. Biomed Pharmacother. 2018;106:1348–1356. doi:10.1016/j.biopha.2018.07.075
26. Pace B, Qian X, Sangerman J, et al. p38 MAP kinase activation mediates γ-globin gene induction in erythroid progenitors. Exp Hematol. 2003;31(11):1089–1096.
27. Haring M, Offermann S, Danker T, Horst I, Peterhansel C, Stam M. Chromatin immunoprecipitation: optimization, quantitative analysis and data normalization. Plant Methods. 2007;24(3):11. doi:10.1186/1746-4811-3-11
28. Jambhekar A, Anastas JN, Shi Y. Histone Lysine Demethylase Inhibitors. Cold Spring Harb Perspect Med. 2017;7(1). doi:10.1101/cshperspect.a026484
29. Lu Z, Ren Y, Zhang M, et al. FLI-06 suppresses proliferation, induces apoptosis and cell cycle arrest by targeting LSD1 and Notch pathway in esophageal squamous cell carcinoma cells. Biomed Pharmacother. 2018;107:1370–1376. doi:10.1016/j.biopha.2018.08.140
30. Forghanifard MM, Taleb S, Abbaszadegan MR. Notch signaling target genes are directly correlated to esophageal squamous cell carcinoma tumorigenesis. Pathol Oncol Res. 2015;21(2):463–467. doi:10.1007/s12253-014-9849-8
31. Lin Y, Kang T, Zhou BP. Doxorubicin enhances Snail/LSD1-mediated PTEN suppression in a PARP1-dependent manner. Cell Cycle. 2014;13(11):1708–1716. doi:10.4161/cc.28619
32. Efferson CL, Winkelmann CT, Ware C, et al. Downregulation of Notch pathway by a gamma-secretase inhibitor attenuates AKT/mammalian target of rapamycin signaling and glucose uptake in an ERBB2 transgenic breast cancer model. Cancer Res. 2010;70(6):2476–2484. doi:10.1158/0008-5472.CAN-09-3114
33. Hales EC, Taub JW, Matherly LH. New insights into Notch1 regulation of the PI3K-AKT-mTOR1 signaling axis: targeted therapy of gamma-secretase inhibitor resistant T-cell acute lymphoblastic leukemia. Cell Signal. 2014;26(1):149–161. doi:10.1016/j.cellsig.2013.09.021
34. Venkatesh V, Nataraj R, Thangaraj GS, et al. Targeting Notch signalling pathway of cancer stem cells. Stem Cell Investig. 2018;5:5. doi:10.21037/sci.2018.02.02
35. Palomero T, Sulis ML, Cortina M, et al. Mutational loss of PTEN induces resistance to NOTCH1 inhibition in T-cell leukemia. Nat Med. 2007;13(10):1203–1210. doi:10.1038/nm1636
 © 2019 The Author(s). This work is published and licensed by Dove Medical Press Limited. The full terms of this license are available at https://www.dovepress.com/terms.php and incorporate the Creative Commons Attribution - Non Commercial (unported, v3.0) License.
By accessing the work you hereby accept the Terms. Non-commercial uses of the work are permitted without any further permission from Dove Medical Press Limited, provided the work is properly attributed. For permission for commercial use of this work, please see paragraphs 4.2 and 5 of our Terms.
© 2019 The Author(s). This work is published and licensed by Dove Medical Press Limited. The full terms of this license are available at https://www.dovepress.com/terms.php and incorporate the Creative Commons Attribution - Non Commercial (unported, v3.0) License.
By accessing the work you hereby accept the Terms. Non-commercial uses of the work are permitted without any further permission from Dove Medical Press Limited, provided the work is properly attributed. For permission for commercial use of this work, please see paragraphs 4.2 and 5 of our Terms.