Back to Journals » OncoTargets and Therapy » Volume 11
Low levels of tumor suppressor candidate 3 predict poor prognosis of patients with hepatocellular carcinoma
Authors Sheng XR, Xing SG, Wang RD , Chen K, Jia WD
Received 6 October 2017
Accepted for publication 9 January 2018
Published 21 February 2018 Volume 2018:11 Pages 909—917
DOI https://doi.org/10.2147/OTT.S153381
Checked for plagiarism Yes
Review by Single anonymous peer review
Peer reviewer comments 3
Editor who approved publication: Dr Carlos E Vigil
Xu-Ren Sheng,1,2,* Song-Ge Xing,2,3,* Run-Dong Wang,2 Kang Chen,2 Wei-Dong Jia1,2
1Department of Liver Surgery, Affiliated Provincial Hospital, Anhui Medical University, 2Anhui Province Key Laboratory of Hepatopancreatobiliary Surgery, 3CAS Key Laboratory of Innate Immunity and Chronic Disease, Innovation Center for Cell Signaling Network, School of Life Sciences, University of Science and Technology of China, Hefei, People’s Republic of China
*These authors contributed equally to this work
Purpose: The tumor suppressor candidate 3 (TUSC3) has been considered to be closely associated with the occurrence, development and invasion of various malignant tumors. However, the expression of TUSC3 in hepatocellular carcinoma (HCC) tissues remains ambiguous. The purpose of this research was to investigate the expression of TUSC3 in HCC tissues and analyze the relationship between TUSC3 levels and clinicopathological characteristics and prognosis of HCC patients.
Materials and methods: Immunohistochemistry was used to detect the expression of TUSC3 in HCC and the corresponding para-cancerous tissues from 92 samples of HCC patients. mRNA and protein expression levels of TUSC3 were evaluated by quantitative real-time polymerase chain reaction (qRT-PCR) and Western blot assays in 25 paired HCC and corresponding adjacent nontumor tissues. Furthermore, statistical analysis was applied to evaluate the correlation between TUSC3 level and the clinicopathological features and prognosis of HCC patients.
Results: Immunohistochemical assay indicated that the expression of TUSC3 was significantly lower in HCC tissues when compared with the corresponding para-cancerous tissues (χ2=11.512, P=0.001). The analysis of clinicopathological characteristics showed that low expression of TUSC3 in HCC tissues was significantly associated with Edmondson grade, Barcelona Clinic Liver Cancer stage and tumor size (P=0.008, 0.009 and 0.020, respectively). Univariate analysis showed that the expression of TUSC3 was strongly correlated with overall survival (OS) and disease-free survival (DFS) after radical surgery in HCC patients (P<0.001, P<0.001, respectively). Multivariate analysis revealed that the TUSC3 level was an independent risk factor for OS and DFS in HCC patients (P=0.001, P<0.001, respectively). Results of qRT-PCR and Western blot assays indicated that the level of TUSC3 in HCC tissues was significantly lower than that in the corresponding adjacent noncancerous tissues (P<0.01, P<0.001).
Conclusion: The expression of TUSC3 in HCC was significantly downregulated and was correlated with tumor progression and prognosis, which could be used as an independent predictor of prognosis in HCC patients.
Keywords: TUSC3, hepatocellular carcinoma, prognosis, immunohistochemistry, overall survival
Introduction
Hepatocellular carcinoma (HCC) is the main type of primary liver cancer, accounting for about 70%–90% of primary liver cancers. HCC is considered as the second leading cause of cancer-related death in men.1 Despite the fact that there are various therapies for HCC, such as surgical resection, portal vein embolization, percutaneous local ablation, transarterial embolization and chemoembolization, and liver transplantation,2,3 its prognosis is still not optimistic. It has been reported that the 5-year survival rate of HCC is only 11%.4 One of the most crucial causes of the gloomy prognosis of HCC is the lack of effective early diagnostic tools. Most patients are diagnosed at an advanced stage and cannot undergo radical surgery. Therefore, it is of great significance to find efficient biological markers with diagnostic values for the early diagnosis and prognosis of HCC.
Tumor suppressor candidate 3 (TUSC3), originally termed as N33, is inspected as a candidate tumor suppressor gene and is located on the human chromosomal band 8p22.5 It is considered to be related to autosomal recessive mental retardation.6–8 In recent years, studies have shown that TUSC3 is closely associated with the occurrence, development and invasion of various malignant tumors such as prostate cancer,9 ovarian cancer,10–12 lung cancer13–16 and colorectal cancer.17 However, the mechanisms of TUSC3 in malignant tumors largely remain unclear and the role of TUSC3 in different tumors is not the same. Few studies have focused on the relationship between TUSC3 and HCC. In view of the lack of sufficient research in this field, our study examined the expression of TUSC3 in HCC tissues and the corresponding para-cancerous tissues from 92 samples of HCC patients by immunohistochemistry analysis. Moreover, quantitative real-time polymerase chain reaction (qRT-PCR) and Western blot assays were applied to detect the mRNA and protein expression of TUSC3 in 25 paired HCC and adjacent nontumor tissues. Finally, we analyzed the relationship between the expression of TUSC3 and clinicopathological features and prognosis of HCC patients.
Materials and methods
Patients and tissue specimens
A total of 92 paraffin samples of HCC patients resected from the Department of Liver Surgery, Affiliated Anhui Provincial Hospital of Anhui Medical University, Hefei, China, from January 2007 to December 2012 were collected for immunohistochemistry analysis. In addition, snap-frozen HCC tissues and the corresponding para-cancerous tissues which were at least 2 cm away from the edge of the tumor from 25 HCC patients with radical HCC resection in Affiliated Anhui Provincial Hospital of Anhui Medical University were obtained for the detection of TUSC3 expression level by qRT-PCR and Western blot assays. All the clinicopathological data such as age, gender, tumor size, vascular invasion (macroscopic or microscopic tumor thrombus of HCC), tumor Edmondson grade, Barcelona Clinic Liver Cancer (BCLC) stage, tumor capsule, serum alpha fetoprotein (sAFP), hepatitis B surface antigen (HBsAg), cirrhosis, Child-Pugh grade and the number of tumors were gathered retrospectively in the medical record room of the hospital. Details of the patients are shown in Table 1. All the specimens were confirmed to be HCC by postoperative pathological examination and all the patients were not treated with radiotherapy, chemotherapy and interventional therapy before operation. The degree of tumor differentiation was determined by Edmondson classification system18 and the nineteenth reference of the BCLC stage criteria.19 The study was followed up for 2–100 months with a median follow-up of 25 months. In addition, the OS time was defined as the time between the date of operation and the date of death or the deadline of follow-up. The disease-free survival (DFS) time was defined as the interval between the date of operation and the date of tumor recurrence which was clearly diagnosed for the first time or the deadline of follow-up. Our study obtained the written informed consent of all patients and the approval of the ethics committee of the clinical laboratory of the Affiliated Anhui Provincial Hospital of Anhui Medical University.
Immunohistochemistry
The paraffin-embedded specimens were sliced into 4 μm thick sections, and the pathological sections were placed in a tissue-drying oven at 60°C for 60 minutes. Then, sections were deparaffinized with xylene and were rehydrated through graded ethanol. Next, slides were steamed in 0.01 M sodium citrate buffer (pH 6.0) at 100°C for 20 minutes, and then were removed from heat and cooled down at room temperature in buffer solution for the sake of antigen retrieval. Sections were incubated with 0.3% hydrogen peroxide to block the activity of endogenous peroxidase for 10 minutes. Tissue slides were incubated at 4°C overnight with rabbit anti-human TUSC3 polyclonal antibody (catalog ID: LS-B13274/68872; LSBio, Seattle, Washington, DC, USA) at a dilution of 1:200, then washed with phosphate-buffered saline (PBS) and incubated with a horseradish peroxidase-conjugated secondary antibody (Zhongshan Golden Bridge Biotechnology, Beijing, China) for 30 minutes. The sections were stained with 3, 3′-diaminobenzidine, counterstained with hematoxylin, dehydrated in grade alcohols and sealed with neutral gum. Immunoreactivity for the expression of TUSC3 was independently assessed by two professional pathologists from the Affiliated Anhui Provincial Hospital of Anhui Medical University. We used a previously published method15 to appraise the semi-quantitative expression of TUSC3 with the following formula: the immunoreactivity score = the staining intensity score × the proportion of positive cells score. The staining intensity score was defined as 0 points (negative), 1 point (weak intensity), 2 points (moderate intensity) and 3 points (strong intensity). The proportion of positive cells score was defined as 0 points (no staining), 1 point (<25%), 2 points (25%–50%), 3 points (50%–75%) and 4 points (>75%). The immunoreactivity scores of the sections which were greater than or equal to 6 points were defined as high expression, otherwise were defined as low expression.
Western blot analysis
Snap-frozen tumor tissues and the corresponding para-carcinomatous tissues were lysed with RIPA buffer (Beyotime Institute of Biotechnology, Shanghai, China) supplemented with protease inhibitor cocktail. The proteins were quantified with bicinchoninic acid protein assay. Equal amounts of the tumor and the corresponding para-carcinomatous specimen proteins were separated by 8% SDS-PAGE and transferred to the polyvinylidene difluoride membranes (EMD Millipore, Billerica, MA, USA). The rabbit anti-human TUSC3 polyclonal antibody (1:500, catalog ID: LS-B13274/68872; LSBio) and β-actin (1:5,000, 66009-1-Ig; Abmart, Arlington, USA) were used to incubate the membranes at 4°C on a shaking table for overnight after blocking with 5% non-fat milk. Next, the membranes were incubated at room temperature with the corresponding secondary antibodies for 2 hours. Signaling was detected by Alpha-EaseFC imaging system (Alpha Innotech, San Leandro, CA, USA). Using the Alpha-EaseFC software, the integrated density value (IDV) of each band was estimated by describing a rectangle outlining the band. Human antibody of β-actin was used as the control, and the ratio of the IDV of TUSC3 to the IDV of β-actin was used for statistical analysis.
qRT-PCR analysis
Total RNA of the 25 pairs of snap-frozen tumors and the corresponding para-carcinomatous tissues was extracted by TRIzol reagent (Thermo Fisher Scientific, Waltham, MA, USA) according to the manufacturer’s protocol, and cDNA was synthesized with the Revert Aid First Strand cDNA Synthesis Kit (Thermo Fisher Scientifc). The mRNA level of TUSC3 was detected by SYBR Green-based RT-PCR with a PikoReal RT-PCR system (Thermo Fisher Scientifc). The PCR amplification was performed according to the following procedure: predenaturation at 94°C for 30 seconds, denaturation at 94°C for 30 seconds, annealing at 60°C for 30 seconds and finally elongation at 72°C for 30 seconds; a total of 40 cycles were performed. We projected specific primers for TUSC3 (Primer Designing Tool, NCBI, Bethesda, MD, USA) as follows: forward: 5′-GAGTTCCAGACGCTCAATCTTC-3′ and reverse: 5′-GCCAGGAGTTCGCCAGTATT-3′. The 18S RNA was used as the internalized control gene, and the sequences used were as follows: forward: 5′-CGGCGACGACCCATTCGAAC-3′ and reverse: 5′-GAATCGAACCCTGATTCCCCGTC-3′. We applied melting-curve analysis to control the purity of PCR products. Each experiment was performed three times with three replicates at each time. The relative expression of TUSC3 mRNA was calculated by 2−ΔΔCt method.
Statistical analysis
SPSS 22.0 software (SPSS Inc, Chicago, IL, USA) and Graph Prism 5.0 software (GraphPad Prism, San Diego, CA, USA) were employed in analyzing the data. Quantitative data involved in this study were expressed as mean ± standard deviation, and the independent Student’s t-test was used for statistical analysis. Chi-square test was performed for qualitative data. The survival curve was drawn by the Kaplan–Meier method, and log-rank test was applied to compare the differences between the survival rates. Cox proportional hazards model was performed for univariate and multivariate analyses of prognostic outcomes. P<0.05 was considered to be statistically significant.
Results
The expression of TUSC3 was downregulated in HCC tissues
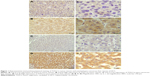
To investigate the expression of TUSC3 in HCC patients, we performed immunohistochemistry analysis to analyze the TUSC3 expression in a retrospective cohort including 92 clinicopathologically characterized HCC patients. Results of immunohistochemistry showed that low expression of TUSC3 was found in 56 (60.9%) HCC tissues and 33 (35.9%) para-cancerous liver tissues, and the difference was statistically significant (χ2=11.512, P<0.05). The positive expression of TUSC3 was mainly observed in the cytoplasm of HCC and para-cancerous liver tissues (Figure 1). Western blot analysis demonstrated that in 19 of the 25 paired tissues (76%), the protein expression of TUSC3 was obviously lower in HCC tissue than the corresponding para-cancerous liver tissues. As shown in Figure 2, the relative protein expression levels of TUSC3 in HCC tissues and the corresponding para-cancerous liver tissues were 0.55±0.34 and 1.07±0.43, respectively (P<0.001). Similarly, qRT-PCR assay revealed that 80% (20/25) of the samples had obviously lower expression, and the relative mRNA expression levels of TUSC3 in HCC tissues and the matched adjacent tissues were 0.64±0.64 and 1.48±1.17, respectively (P<0.01), as shown in Figure 3.
Low expression of TUSC3 in HCC tissues was closely related to Edmondson grade, BCLC stage and tumor size
Next, we analyzed the correlation between the expression of TUSC3 and clinicopathological data of HCC. Results indicated that low expression of TUSC3 in HCC tissues was closely associated with Edmondson grade, BCLC stage and tumor size, and the difference was statistically significant (P=0.008, P=0.009, P=0.020, respectively). However, no statistically significant correlation was found between the TUSC3 level and age, gender, tumor capsule, vascular invasion, sAFP, HBsAg, cirrhosis, Child-Pugh grading and tumor number, and the difference was not statistically significant (P>0.05) (Table 1). These abovementioned results suggested that the low expression of TUSC3 might be related to the malignant process of HCC.
Low TUSC3 levels predicted poor prognosis of patients with HCC
Then we analyzed the relationship between the expression of TUSC3 and the prognosis of HCC patients by collecting the prognosis data for all patients. The patients in our study were followed up for 2–100 months and the median follow-up time was 25 months. Kaplan–Meier method was performed for survival analysis, and the results showed that patients with lower TUSC3 expression had a shorter OS time (17 months, 95% confidence interval [CI]: 13.010–20.990) than those with higher TUSC3 expression (36 months, 95% CI: 25.547–46.453; P<0.001). Analogously, the DFS survival curves revealed that the patients with lower TUSC3 expression had a shorter DFS time when compared with those with higher TUSC3 expression (9 months, 95% CI: 6.529–11.471 vs 17 months, 95% CI: 10.101–23.899) (Figure 4). Moreover, univariate analysis indicated that the TUSC3 expression, Edmondson grade, BCLC stage, tumor capsule, tumor size and vascular invasion were closely correlated with the OS and DFS of HCC patients (Table 2). Multivariate analysis showed that the TUSC3 level, tumor capsule and vascular invasion were independent risk factors for OS and DFS in postoperative patients with HCC (Table 3).
Discussion
The protein encoded by TUSC3 is a subunit in the oligosaccharyltransferase (OST) complex that participates in the N-linked glycosylation of the protein folding process.9,20 Abnormal expression of TUSC3 may change the process of N-linked glycosylation, and the abnormity of N-linked glycosylation which participates in biological processes, such as intercellular or cell–matrix communications, plays a crucial role in the occurrence and progression of tumor.21–24 Therefore, more and more studies have explored the link between TUSC3 and malignant tumors. For instance, Horak et al have reported that TUSC3 expression is significantly associated with promoter hyper-methylation as well as downregulation in advanced prostate cancer patients and indicated that deletion of TUSC3 in prostate cancer cells may increase the proliferation and invasion of cancer cells. Therefore, TUSC3 may act as a tumor suppressor gene in prostate cancer.9 Analogously, it has been reported that the expression of TUSC3 in ovarian cancer is distinctly decreased due to the methylation of the TUSC3 promoter region through the study of ovarian cancer cell lines and tumor samples, and the absence of TUSC3 promotes the proliferation and migration of ovarian cancer cells, which may be related to abnormal N-linked glycosylation.11,12 In addition, another study has shown that the expression of TUSC3 is markedly lower in small-cell lung cancer than that in the normal control, and the low expression of TUSC3 is associated with poorly differentiated lung cancer.15 It is also suggested that TUCS3 may be a predictor of lymph node metastasis in patients with lung cancer by analyzing the correlation between TUSC3 expression and lymph node metastasis.15 In contrast, there are also studies of the opposite views about the tumorigenic ability of TUSC3. For example, Gu et al have found that TUSC3 is highly expressed in non-small-cell lung cancer and the overexpression of TUSC3 can enhance cell proliferation, migration and tumor growth ability of human non-small-cell lung cancer cells, which means that TUSC3 plays a carcinogenic role in non-small-cell lung cancer.14 Moreover, another study has revealed that TUSC3 expression is upregulated in colorectal cancer specimens and its overexpression in colorectal cancer cells also increases the abilities of proliferation, migration and tumorigenesis.17 Based on previous studies, our research has found that TUSC3 played disparate roles in different tumors. However, the expression and clinical significance of TUSC3 in HCC remain uncertain.
In this study, through immune-histochemical experiments, we found that the expression of TUSC3 in HCC was obviously lower than that in the corresponding para-cancerous tissues. Low expression of TUSC3 in HCC patients was significantly associated with adverse clinical features including Edmondson grade, BCLC stage and tumor size. Furthermore, results of Western blot and qRT-PCR indicated that both the protein and mRNA expression of TUSC3 were significantly downregulated in HCC tissues, which were consistent with the immune-histochemical results. We hypothesized that downregulation of TUSC3 in HCC might be related to the methylation of promoter regions; however, the detailed mechanisms need to be further studied. In brief, the abovementioned results suggested that the expression of TUSC3 was low in HCC and its downregulation might be associated with HCC growth, invasion and tumor differentiation.
The specific regulatory mechanism of TUSC3 in tumors is not yet clear. Horak et al have suggested that the loss of TUSC3 leads to increased N-glycosylation and alleviation of endoplasmic reticulum stress which can enhance the ability of tumorigenesis in prostate cancer cells.9 They have also reported that TUSC3 plays an important role in tumor inhibition through distinct regulatory mechanisms including prevention of the epithelial-to-mesenchymal transition and modulation of the endoplasmic reticulum stress response in ovarian cancer cells.10 Prior studies have found that TUSC3 promotes autophagy in human non-small-cell lung cancer cells by activating the Wnt/β-catenin signaling pathway, and the tumorigenic role of TUSC3 in non-small-cell lung cancer may be related to Hedgehog signaling pathway.13,14 It has also been reported that TUSC3 acts as an oncogene in colorectal cancer by inducing epithelial-to-mesenchymal transition and potentially regulating the MAPK, PI3K/Akt and Wnt/β-catenin signaling pathways.17 Several studies have also demonstrated that TUSC3 acts as a tumor suppressor in glioblastoma by inhibiting the activity of the Akt signaling pathway, and its role of a target protein is closely related to the formation and progression of glioblastoma.25–27 Based on the abovementioned studies, we can conclude that the regulatory mechanisms of TUSC3 in tumors may include many aspects. Unfortunately, we failed to elaborate on the molecular mechanism of TUSC3 in the formation and progression of HCC in this study. We speculated that TUSC3 played a role of tumor suppressor by regulating endoplasmic reticulum stress response in HCC, and the specific molecular mechanism needs to be further studied as a direction of our further work.
Previous studies have demonstrated the prognostic value of TUSC3 in other malignancies. Pils et al have observed that TUSC3 promoter methylation is significantly related to shorter progression-free and OS rates.12 It has been reported that patients with negative expression of TUSC3 have obviously shorter OS in esophageal squamous cell carcinoma.28 Nevertheless, Duppel et al have found that TUSC3 methylation is markedly associated with a longer OS time in lung cancer.16 Our results obtained by the Kaplan–Meier method indicated that patients with low expression of TUSC3 had shorter OS and DFS than those with high expression of TUSC3. In addition, we concluded that TUSC3 was an independent prognostic factor for HCC patients after surgery by using the Cox proportional hazards model.
In summary, we revealed for the first time that the expression of TUSC3 was downregulated in HCC and its low expression was associated with adverse clinical features and poor OS and DFS in HCC patients. TUSC3 might play a crucial role in the early diagnosis and prognosis prediction of HCC which was also important in clinical studies. However, the exact molecular mechanisms by which TUSC3 promotes HCC progression remain unclear and need to be further studied.
Acknowledgment
This study was supported by Anhui Science and Technology Public Relations Projects (No 1704a0802150).
Disclosure
The authors report no conflicts of interest in this work.
References
Torre LA, Siegel RL, Ward EM, Jemal A. Global cancer incidence and mortality rates and trends – an update. Cancer Epidemiol Biomarkers Prev. 2016;25(1):16–27. | ||
Crocetti L, Bargellini I, Cioni R. Loco-regional treatment of HCC: current status. Clin Radiol. 2017;72(8):626–635. | ||
Rahbari NN, Mehrabi A, Mollberg NM, et al. Hepatocellular carcinoma: current management and perspectives for the future. Ann Surg. 2011;253(3):453–469. | ||
Blechacz B, Mishra L. Hepatocellular carcinoma biology. Recent results in cancer research. Fortschritte der Krebsforschung. Progres Dans Les Recherches Sur Le Cancer. 2013;190:1–20. | ||
MacGrogan D, Levy A, Bova GS, Isaacs WB, Bookstein R. Structure and methylation-associated silencing of a gene within a homozygously deleted region of human chromosome band 8p22. Genomics. 1996;35(1):55–65. | ||
Garshasbi M, Hadavi V, Habibi H, et al. A defect in the TUSC3 gene is associated with autosomal recessive mental retardation. Am J Hum Genet. 2008;82(5):1158–1164. | ||
Loddo S, Parisi V, Doccini V, et al. Homozygous deletion in TUSC3 causing syndromic intellectual disability: a new patient. Am J Med Genet A. 2013;161A(8):2084–2087. | ||
Garshasbi M, Kahrizi K, Hosseini M, et al. A novel nonsense mutation in TUSC3 is responsible for non-syndromic autosomal recessive mental retardation in a consanguineous Iranian family. Am J Med Genet A. 2011;155A(8):1976–1980. | ||
Horak P, Tomasich E, Vanhara P, et al. TUSC3 loss alters the ER stress response and accelerates prostate cancer growth in vivo. Sci Rep. 2014;4:3739. | ||
Kratochvilova K, Horak P, Esner M, et al. Tumor suppressor candidate 3 (TUSC3) prevents the epithelial-to-mesenchymal transition and inhibits tumor growth by modulating the endoplasmic reticulum stress response in ovarian cancer cells. Int J Cancer. 2015;137(6):1330–1340. | ||
Vanhara P, Horak P, Pils D, et al. Loss of the oligosaccharyl transferase subunit TUSC3 promotes proliferation and migration of ovarian cancer cells. Int J Oncol. 2013;42(4):1383–1389. | ||
Pils D, Horak P, Vanhara P, et al. Methylation status of TUSC3 is a prognostic factor in ovarian cancer. Cancer. 2013;119(5):946–954. | ||
Peng Y, Cao J, Yao XY, et al. TUSC3 induces autophagy in human non-small cell lung cancer cells through Wnt/beta-catenin signaling. Oncotarget. 2017;8(32):52960–52974. | ||
Gu Y, Pei X, Ren Y, et al. Oncogenic function of TUSC3 in non-small cell lung cancer is associated with Hedgehog signalling pathway. Biochim Biophys Acta. 2017;1863(7):1749–1760. | ||
Yu X, Zhang K, Liu F, et al. Tumor suppressor candidate 3 as a novel predictor for lymph node metastasis in lung cancer patients. Oncol Lett. 2016;12(6):5099–5105. | ||
Duppel U, Woenckhaus M, Schulz C, Merk J, Dietmaier W. Quantitative detection of TUSC3 promoter methylation – a potential biomarker for prognosis in lung cancer. Oncol Lett. 2016;12(4):3004–3012. | ||
Gu Y, Wang Q, Guo K, et al. TUSC3 promotes colorectal cancer progression and epithelial-mesenchymal transition (EMT) through WNT/beta-catenin and MAPK signalling. J Pathol. 2016;239(1):60–71. | ||
Edmondson HA, Steiner PE. Primary carcinoma of the liver: a study of 100 cases among 48,900 necropsies. Cancer. 1954;7(3):462–503. | ||
Forner A, Reig ME, de Lope CR, Bruix J. Current strategy for staging and treatment: the BCLC update and future prospects. Semin Liver Dis. 2010;30(1):61–74. | ||
Mohorko E, Owen RL, Malojcic G, Brozzo MS, Aebi M, Glockshuber R. Structural basis of substrate specificity of human oligosaccharyl transferase subunit N33/Tusc3 and its role in regulating protein N-glycosylation. Structure. 2014;22(4):590–601. | ||
Helenius A, Aebi M. Intracellular functions of N-linked glycans. Science. 2001;291(5512):2364–2369. | ||
Oliveira-Ferrer L, Legler K, Milde-Langosch K. Role of protein glycosylation in cancer metastasis. Semin Cancer Biol. 2017;44:141–152. | ||
Li JH, Huang W, Lin P, et al. N-linked glycosylation at Asn152 on CD147 affects protein folding and stability: promoting tumour metastasis in hepatocellular carcinoma. Sci Rep. 2016;6:35210. | ||
Hakomori S. Glycosylation defining cancer malignancy: new wine in an old bottle. Proc Natl Acad Sci U S A. 2002;99(16):10231–10233. | ||
Jiang Z, Guo M, Zhang X, et al. TUSC3 suppresses glioblastoma development by inhibiting Akt signaling. Tumour Biol. 2016;37(9):12039–12047. | ||
Liang Q, Wang K, Wang B, Cai Q. HCMV-encoded miR-UL112-3p promotes glioblastoma progression via tumour suppressor candidate 3. Sci Rep. 2017;7:44705. | ||
Cheng ZX, Yin WB, Wang ZY. MicroRNA-132 induces temozolomide resistance and promotes the formation of cancer stem cell phenotypes by targeting tumor suppressor candidate 3 in glioblastoma. Int J Mol Med. 2017;40(5):1307–1314. | ||
Yu X, Zhang J, Zhong H, et al. Decreased tumor suppressor candidate 3 predicts poor prognosis of patients with esophageal squamous cell carcinoma. Int J Med Sci. 2016;13(12):963–969. |
 © 2018 The Author(s). This work is published and licensed by Dove Medical Press Limited. The full terms of this license are available at https://www.dovepress.com/terms.php and incorporate the Creative Commons Attribution - Non Commercial (unported, v3.0) License.
By accessing the work you hereby accept the Terms. Non-commercial uses of the work are permitted without any further permission from Dove Medical Press Limited, provided the work is properly attributed. For permission for commercial use of this work, please see paragraphs 4.2 and 5 of our Terms.
© 2018 The Author(s). This work is published and licensed by Dove Medical Press Limited. The full terms of this license are available at https://www.dovepress.com/terms.php and incorporate the Creative Commons Attribution - Non Commercial (unported, v3.0) License.
By accessing the work you hereby accept the Terms. Non-commercial uses of the work are permitted without any further permission from Dove Medical Press Limited, provided the work is properly attributed. For permission for commercial use of this work, please see paragraphs 4.2 and 5 of our Terms.