Back to Journals » Cancer Management and Research » Volume 14
Low Expression of ZNF154 is Related to Poor Prognosis in Gastric Cancer
Authors He J, Huang J, Tang G, Wang P, He M, Wei S
Received 18 September 2021
Accepted for publication 3 January 2022
Published 17 February 2022 Volume 2022:14 Pages 659—672
DOI https://doi.org/10.2147/CMAR.S340053
Checked for plagiarism Yes
Review by Single anonymous peer review
Peer reviewer comments 2
Editor who approved publication: Professor Yong Teng
Jinsong He,1,* Jing Huang,2,* Guo Tang,1 Pan Wang,1 Ming He,3 Shoujiang Wei1
1Department of Gastroenterology, Affiliated Hospital of North Sichuan Medical College, Nanchong, 637000, People’s Republic of China; 2Lung Cancer Institute, Affiliated Hospital of North Sichuan Medical College, Nanchong, 637000, People’s Republic of China; 3Graduate School of North Sichuan Medical College, Nanchong, 637000, People’s Republic of China
*These authors contributed equally to this work
Correspondence: Shoujiang Wei, Email [email protected]
Introduction: Zinc finger protein 154 (ZNF154) has been identified as a tumor suppressor gene in multiple carcinomas. Lymph node (LN) metastasis is one of the most intensively negative factor of gastric cancer (GC) prognosis. However, the potential mechanisms of ZNF154-mediated LN metastasis are not elucidated. This study aimed to investigate the role of ZNF154 in LN metastasis of GC and their underlying mechanisms through in vitro and in vivo experiments.
Methods: Antitumor effect was measured by growth inhibition by cell counting kit-8 (CCK-8) and colony formation assay. Cell cycle distribution and apoptosis were evaluated by flow cytometry. Cell migration and invasion were measured by wound healing and transwell invasion assays, respectively. The expression levels of proteins were analyzed by Western blot. Xenograft models were used for validation in vivo.
Results: Our research showed that ZNF154 was down-regulated in 81.43% (57 of 70) of GC tissues compared with 58.6% of paired non-tumor tissues from patients, ZNF154 was down-regulated in 100% (7 of 7) of GC cell lines, up-regulated expression of ZNF154 in MGC-803 GC cells reduced cell proliferation, viability, migration and invasion, and enhanced cell apoptosis and arrested cell cycle in G2 phase, and suppressed tumorigenicity of MGC-803 cells in mice. Furthermore, up-regulated expression of ZNF154 mRNA reduced the expression of B-cell lymphoma-2 (Bcl-2), matrix metalloproteinase 2 (MMP-1), hepatocyte growth factor (HGF), vascular endothelial growth factor-A/C (VEGF-A/C).
Conclusion: ZNF154 inhibited LN metastasis of GC cells by suppressing several biological events of GC cells. ZNF154 was a tumor suppressor gene that is a promising target for blocking nodal involvement in GC.
Keywords: ZNF154, gastric cancer, lymph node, metastasis, prognosis
Introduction
Gastric cancer (GC) is one of the most common malignancies worldwide. Nearly 40% of the global GC cases occur in China, 80% of which are diagnosed at an advanced stage, and the 5-year overall survival (OS) rate is less than 50%. LN metastasis is recognized as an independent prognostic factor for GC, the occurrence of LN metastasis will greatly increase the risk of postoperative recurrence and shorten postoperative survival time. Zinc finger proteins (ZFPs) are the largest family of transcription factors in the human genome, and bind to gene promoters via zinc finger domains to activate or repress gene expression. Unfortunately, the role of ZFPs in the occurrence and development of GC has not been fully clarified.
The zinc finger protein 154 (ZNF154) gene have been reported as a tumor suppressor gene in the zinc finger protein family.1 Compelling evidence has shown that ectopic expression of the ZNF154 gene would suppress tumor cell growth, migration, invasion, and colony formation in nasopharyngeal carcinoma.2 Studies have shown that the count of methylated CpG sites in the ZNF154 DNA promoter was positively associated with longer survival in respectable pancreatic cancer.1 Hypermethylation of ZNF154 leads to inhibition of gene expression inhibition and loss of function, which contribute to cancer development and poor patient outcomes.3,4 In addition, researchers recently revealed a novel molecular mechanism. They found that ZNF154 could suppress invasion and metastasis of nasopharyngeal carcinoma (NPC) by inhibiting the epithelial-mesenchymal transition (EMT) through Wnt/β-catenin signaling pathway.2
We know that lymph node metastasis is one of the most intensively negative factor of GC prognosis. However, the mechanism of lymph node metastasis has not been fully elucidated. Thus, it is essential to explore the potential mechanisms of ZNF154-mediated lymph node (LN) metastasis in the progression of GC to further evaluate the feasibility of targeting therapy using the ZNF154 gene. In the present study, we aimed to investigate the role of ZNF154 in LN metastasis of GC and their underlying mechanisms through in vitro and in vivo experiments.
Materials and Methods
Patient Cohort and Database
Seventy fresh tumor and adjacent non-tumor tissue samples were obtained from GC patients who underwent curative gastrectomy between November 2015 and September 2019 at the Affiliated Hospital of North Sichuan Medical College. All surgical specimens were assessed pathologically in accordance with the International Union Against Cancer TNM (UICC) classification for GC. Patients were followed up regularly in the clinic. This study was conducted in accordance with the Declaration of Helsinki, and all related procedures were approved by the ethics committee of North Sichuan Medical College. All included patients provided written informed consent.
GC Surgical Procedure
Curative resection was defined as the complete absence of grossly visible tumor tissues and metastatic lymph nodes after resection, with pathologically negative resection margins. Primary tumors were resected en bloc with limited or extended lymphadenectomy.
Immunohistochemistry
Formalin-fixed, paraffin-embedded cancer tissue specimens from 70 GC patients and 70 paired adjacent non-tumor tissue samples were analyzed by immunohistochemical staining to detect ZNF154 protein expression. Specimens were incubated with monoclonal antibody against ZNF154 (1:500; bs-13723R-Cy3; Bioss, Beijing, China). The nuclear expression of ZNF154 was assessed according to the average intensity of positively stained cells (“-” was none, “+” was weak, “++” was intermediate, “+++” was strong). More than “++” were classified into positive staining group.5
Cell Culture
Seven GC cell lines (AGS, Kato-III, SUN-1, SGC-7901, HGC-27, MGC-803, and BGC-823) were purchased from the Type Culture Collection of the Chinese Academy of Sciences (Shanghai, China). Human normal gastric mucosa cell line GES-1 lines were purchased from Biowit Technologies Corporation (Shenzhen, China). Cells were cultured in RPMI-1640 medium (Thermo Electron Corporation, Beijing, China) supplemented with 10% fetal bovine serum (Life Tech, Mulgrave Vic, Australia) at 37°C atmosphere with 5% CO2.
Quantitative Real-Time PCR (RT-qPCR)
The total RNA was isolated using TRIzol® reagent (Invitrogen, Carlsbad, CA, USA) and cDNA was synthesized using Transcriptor Reverse Transcriptase (Roche Diagnostics, Shanghai, China) followed by quantitative reverse transcription q-PCR according to the manufacturer’s instructions.
 |
|
Construction Vectors and Transfection
A mammalian expression vector (pCMV6-AC-GFP) (Biovector, USA) was used to generate the ZNF154 upregulated expression vector (pCMV6-AC-GFP-ZNF154) or empty vector (pCMV6-AC-GFP). LipofectamineTM 3000 transfection reagent (Invitrogen) was used to transfect the vectors into MGC-803 cells according to the manufacturer’s instructions.6
Colony Formation Assay
Colony formation detection was performed as previously described by Yu et al.7 G418 sulphate (Merck, Darmstadt, Germany) was used in colony formation experiments with transfected MGC-803 cells. The assay was performed three times.
Cell Viability
Cell Counting Kit-8 (CCK8) assays were performed to determine the viability of transfected MGC-803 cells, the procedure was described by Germack and Dickenson.8
Cell-Based Scratch Assay
A scratch wound assay was applied to assess the migration ability of transfected MGC-803 cells as described by Sung et al.9
Transwell Tumor Cell Invasion Assay
Matrigel™ (Sigma-Aldrich) invasion experiments were performed as described by Zhou et al.10 Cell invasion assays were applied to detect the invasion ability of transfected MGC-803 cells. All experiments were performed in triplicate.
Measurement of Apoptosis
Apoptosis was measured using Annexin V and propidium iodide dual staining following the manufacturer’s instructions by method previously described by Das et al.11
Cell Cycle Distribution Analysis
Flow cytometry was conducted to determine the cell cycle-phase distribution, transfected MGC-803 cells were harvested and the experimental procedures were performed by a previously reported method.7
In vivo Tumorigenicity
For the tumorigenesis ability test, MGC-803 cells (1 × 106 cells in 0.1 mL phosphate-buffered saline) transfected with pCMV6-AC-GFP–ZNF154 or pCMV6-AC-GFP empty vector were injected subcutaneously into the dorsal flank of each 3-week-old male nude mice, separately. Tumor size was estimated every 2 or 3 days until 3 weeks by measuring the longest diameter of the tumor. Care of animals and all experimental procedures were performed according to the Animal Care Facility and were approved by the Animal Ethics Committee of Affiliated Hospital of North Sichuan Medical College.
Statistical Analysis
The results are expressed as mean ± standard deviation (SD). The Mann–Whitney U-test was performed to compare the difference in ZNF154 protein expression between tumor and adjacent non-tumor tissues. The difference in tumor growth rate between the two groups of nude mice was determined by repeated-measures analysis of variance. The median overall survival (OS) was determined with the Kaplan–Meier method, and Log rank test was used to determine significance. Multivariate analysis of the OS was performed with the Cox proportional hazards model. Hazard ratios (HRs) and 95% confidence intervals (CIs) were generated. The χ2 test was used for comparison of patient characteristics and distributions of ZNF154 expression and covariates by vital status. The likelihood test in multinomial logistic regression was adopted in terms of different categories to quantify their discriminatory ability. With the cut-point survival analysis, the optimal cutoffs for continuous variables were identified.12 Significance was defined as P < 0.05. All statistical analyses were performed with SPSS 23.0 software.
Results
Clinical Patient Series
ZNF154 is Significantly Downregulated in Human GC Tissues
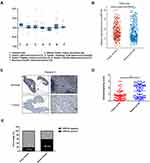
To investigate the expression pattern of ZNF154 in human GC tissues, two data sets were used (https://tcga-data.nci.nih.gov/)(www.oncomine.com). Data were collected and analyzed using R3.1.2 software. We found that the ZNF154 mRNA level was significantly decreased in all types of GC, including diffuse gastric adenocarcinoma, gastric adenocarcinoma, gastric intestinal-type adenocarcinoma, gastric papillary adenocarcinoma, gastric tubular adenocarcinoma, and mucinous gastric adenocarcinoma compared with that in normal colorectal tissues (Figure 1A).
Furthermore, fresh tumor and adjacent normal tissue samples were collected from 70 GC patients. Median follow-up from the time of diagnosis was 44 (range, 4–100) months. The clinical data and patient characteristics are shown in Table 1. The expression intensity was classified as none (−) in 39 (55.7%) patients, weak (+) in 18 (25.7%) patients, intermediate (++) in 11 (15.7%) patients, and strong (+++) in 2 (2.9%) patients. The results of ZNF154 mRNA expression were consistent with the protein expression levers (mean (SD), 0.201 (0.064) vs 0.446 (0.105); P < 0.001, Figure 1B). ZNF154 protein in GC and the paired adjacent non-tumor tissues were expressed mainly in the nucleus through immunohistochemical staining (Figure 1C and D). Thus, only 13 (18.6%) patients showed positive expression of ZNF154 protein. Moreover, ZNF154 was positively expressed in 41 (58.6%) paired adjacent non-tumour tissues. compared with paired adjacent non-tumor tissues, ZNF154 protein expression was downregulated significantly in GC tissues (χ2 = 23.635, P < 0.001, Figure 1E).
 |
Table 1 Patient Demographics |
Downregulation of ZNF154 is Associated with Lymph Node Metastasis and Poor Prognosis in GC Patients
Table 2 summarizes the results of univariate and multivariate survival analysis of the 70 GC patients. Median OS of all GC patients was 43 months. Further prognostic analysis revealed that patients who positively expressed ZNF154 protein displayed better survival than those with negative expression (P < 0.001) (Figure 2A). ZNF154 protein expression in GC tissues was identified as an independent predictor associated with poor OS (HR = 3.263, P = 0.031), along with pN stage and tumor size. Therefore, positive ZNF154 expression in tumor samples indicated a tendency of comparatively longer OS in GC patients.
 |
Table 2 Survival Analysis of 70 GC Patients |
We further explored the relationship between the expression of ZNF154 and LN metastasis and survival status, we reviewed TCGA with assistance of the Clinical Bioinformatics database. The Sankey diagram of TCGA showed the high and low expression distribution trend of the ZNF154 gene in different N stages and survival status in GC samples (Figure 2B). We investigated the correlations of ZNF154 protein expression (immunohistochemical staining) and clinicopathological parameters of GC patients in Chinese database. The results are shown in Table 3. The number of lymph node metastases (pN stage) was validated as more relevant to ZNF154 expression (P = 0.002). As the number of lymph node metastases increased, the positive rate of ZNF154 expression in tumor tissues of GC patients was reduced (40.0% for pN1 patients, 37.5% for pN2 patients, 33.3% for N3a patients, and 0% for N3b patients; P = 0.002).
 |
Table 3 Correlation Analyses Between ZNF154 Protein Expression and Clinicopathological Characteristics |
ZNF154 in GC Cell Lines
ZNF154 Enhances GC Cell Proliferation and Apoptosis and Modulates the Cell Cycle Distribution
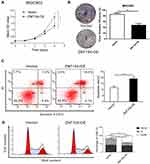
To investigate the effects of ZNF154 upregulated expression on the growth and viability of GC cells, we utilized colony formation and CCK8 assays. The CCK8 assay revealed that the mean OD values of MGC-803-pCMV6-AC-GFP-ZNF154 cells (MGC803-ZNF154) were lower than that of MGC-803-pCMV6-AC-GFP empty vector cells (MGC803-vector) between the 24 h and 96 h time-points (Figure 3A). Meanwhile, colony formation showed that MGC-803-ZNF154 cells would form fewer colonies compared with MGC-803-vector cells (P = 0.003) (Figure 3B). Compared with the apoptosis of MGC803-vector cells, apoptosis rates of MGC803-ZNF154 cells were significantly increased. The mean cell apoptosis rates were 5.3% and 9.6% for MGC803-vector and MGC803-ZNF154 cells, respectively (Figure 3C), indicating that overexpression of ZNF154 enhanced the apoptosis of MGC-803 cells (P < 0.001). Flow cytometry was conducted to determine whether ZNF154 was associated with delay in MGC-803 cell cycle. Compared with the MGC-803-vector cell line, the MGC803-ZNF154 cell line presented higher cell counts in G2 phase and lower cell counts in S phase at 72 h. The mean diploid ratios of G2 phase cell counts of two cancer cell lines (MGC803-ZNF154 and MGC-803-vector) were 42.48% and 17.44%, respectively (P = 0.026) (Figure 3D). The results clearly suggest that the overexpression of ZNF154 can inhibit cell synthesis of MGC-803 cancer cells.
ZNF154 Inhibits GC Cell Migration and Invasion
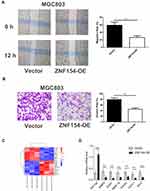
A scratch assay was performed to observe the function of ZNF154 in GC cell migration. As shown in Figure 4A, the groups transfected with ZNF154 (MGC803-ZNF154 cells) could significantly inhibit cell migration compared with that in the empty control (MGC803-vector). The inhibition rates of migration were 66.33% for MGC-803 cells at 24 h (Figure 4B) Taken together, our findings show ZNF154 inhibits migratory and invasive behaviors in GC cells.
ZNF154 Modulates the Biological Characteristic Genes of GC Cells
The expression level of ZNF154 in MGC803-ZNF154 cells was higher than that in MGC-803-vector cells by qPCR (Supplemental Figure 1). We further explored the potentially molecular mechanism of ZNF154 inhibition of malignant biological characteristics of GC cells. As shown in Figure 4C and D, the mRNA levels of generally characterized biomarkers involved in human tumorigenesis were different between MGC803-ZNF154 and MGC803-vector cells. The results found that ZNF154 could significantly affect the gene expression of proliferation (EGR1, FGFR1, P < 0.001), invasion (MMP-9, MMP-19, P < 0.001), lymph angiogenesis (CXCL1, P < 0.001) and anti-apoptosis (Bcl-2, P = 0.0004) of GC cells.
Experiments Involving Nude Mice
ZNF154 Inhibits Tumorigenesis in vivo
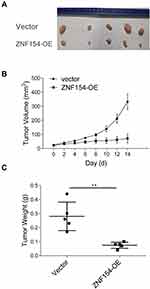
To demonstrate the above in vitro results, an in vivo xenograft model was used. The tumor growth curve of MGC803-ZNF154 and MGC803-vector cells injected subcutaneously in nude mice is shown in Figure 5C. Three weeks after cell implantation, the mean tumor size was significantly smaller in MGC803-ZNF154-injected nude mice than MGC-803-vector cell (27.2 vs 47.8 mm, P < 0.001), indicating that ZNF154 functioned as a tumor suppressor gene in GC.
Discussion
Zinc finger proteins are the largest transcription factor family in the human genome. The diverse combinations and functions of zinc finger motifs make zinc finger proteins play a variety of roles in biological processes, including development, differentiation, metabolism, and autophagy. Over the past few decades, evolving evidence has revealed the potential roles of zinc finger proteins in cancer progression. However, because zinc finger proteins work differently in different cancer types or even in the same cancer type under different types of stress.13–16 The underlying mechanisms in cancer progression have not been clarified yet.
In addition to cancer promotion, several ZNFs have been found to function as tumor suppressors.17–21 As a member of the zinc finger protein family, expression of ZNF154 has been shown to be downregulated in many kinds of human malignancies, due to DNA promoter methylation-induced silencing. DNA promoter methylation has been reported to be the crucial cause of ZNF154 downregulation in oncogenesis and tumor progression of a wide variety of cancers.22–27 Hypermethylation of ZNF154 has been reported in breast,22 lung,23 hepatocellular,24 ovarian,25 renal,26 and prostate cancer.27 Previous analysis of DNA methylation data in TCGA cohorts has also revealed that the ZNF154 transcription start site (TSS) is hypermethylated in 15 different solid epithelial tumor types compared with baseline methylation in normal tissue.28
In addition, a further study indicated that ZNF154 hypermethylation may represent a pan-cancer biomarker for solid tumors and could potentially be used to diagnose tumors through circulating tumor DNA.3 In the present study, the positive rate of ZNF154 expression in GC tissues was also identified to correlate with a significantly increased risk for increased lymph node metastatic count. Therefore, it was proven that ZNF154 ought to be in favor of prognostic improvement of GC by inhibiting nodal involvement.
Lymph node metastasis is the most relevant prognostic indicator in gastric cancer. Meticulous examination of lymph node metastasis is essential in accurately predicting the prognosis in patients with gastric cancer. Occurrence of LN metastasis from GC includes a series of abnormal molecular events in GC cells. Several prerequisites for inducing LN metastasis of cancer cells include proliferation, migration, invasion, apoptosis, cell cycle distribution, and lymph angiogenesis.29,30 A recent study demonstrated that ZNF154 is commonly downregulated or silenced by promoter methylation in nasopharyngeal carcinoma.2 The authors further elucidated that ZNF154 can suppress nasopharyngeal carcinoma cell invasion and metastasis through inhibiting Wnt/β-catenin signaling pathway activation and suppressing the EMT. Moreover, hypermethylation of the ZNF154 promoter is associated with significantly poorer survival in patients with locally advanced nasopharyngeal carcinoma, who are at high risk of metastasis.2 A recent study also showed the effects of ZNF154 on cell viability and gene expression in vitro.1 In the present study, ZNF154 upregulation revealed obvious effects on the growth and viability of MGC-803 GC cells, indicating that ZNF154 silencing should be considered as a relevant factor for GC growth. ZNF154 re-expression decreased the mRNA level of HGF, a specifically proliferative gene, in MGC-803 cells, indicating a potential molecular mechanism of ZNF154-mediated inhibition of GC growth. Furthermore, analysis of tumorigenesis in vivo also confirmed that ZNF154 functioned specifically in growth inhibition. ZNF154 was also shown to be capable of inhibiting migration and invasion of MGC-803 GC cells by the scratch assay and the Matrigel model, indicating that ZNF154 silencing might contribute to cellular infiltration in GC. Upregulated ZNF154 expression inhibition the MMP-2 mRNA level in the MGC-803-ZNF154 cells was illuminated in this study, indicating ZNF154 decreased the invasiveness of GC cells.
ZNF154 encodes a zinc-finger protein. Zinc-finger proteins are, among other functions, involved in transcriptional activation and regulation of apoptosis. However, upregulated expression of ZNF154 was demonstrated to enhance the apoptosis and inhibit cell synthesis of MGC-803 cancer cells by arresting cells in G1 phase in this study. Expression of the anti-apoptotic gene Bcl-2 was inhibited in MGC-803-ZNF154 cells in this study. Therefore, ZNF154 could promote apoptosis of GC cells.
Lastly, downregulated expression of VEGF-C in MGC-803-ZNF154 cells provided a clue that ZNF154 might suppress lymph angiogenesis in GC, which should be considered as a crucial condition for ZNF154-mediated inhibition of lymphatic invasion of GC cells. Although VEGF-A mainly contributes to angiogenesis, it has recently been shown that VEGF-A could induce phosphorylation of VEGFR-2 to promote lymph angiogenesis and metastasis in oral squamous cell carcinoma.28 Therefore, ZNF154 may restrain lymph angiogenesis in GC by regulating the VEGF-A/C-VEGFR-2/3 axis, which should be deemed as a key molecular mechanism of ZNF154-mediated inhibition of LN metastasis of GC cells. Nevertheless, the expression regulation of ZNF154 is still worthy of further exploration. In the future, we will continue to explore how to re-express ZNF154 in GC cells.
Conclusions
In conclusion, ZNF154 inhibited LN metastasis of GC cells by suppressing several biological events of GC cells, including proliferation, viability, invasion, migration, anti-apoptosis mechanisms, and tumor lymph angiogenesis. The present study represents a novel exploration of mechanism of biomarker variable evaluation of the LN metastasis of GC prior to its clinical application. Our findings suggested that ZNF154 was a tumor suppressor gene that is a promising target for blocking nodal involvement in GC.
Data Sharing Statement
The data used to support the findings of this study are available from the corresponding author upon request.
Ethics Approval and Consent to Participate
This research was approved by the Ethics Committee of Affiliated Hospital of North Sichuan Medical College, and oral and written inform consents were obtained from all patients before enrolling in the research program. The in vivo assay using nude mice was approved by the Institutional Animal Care and Use Committee of Affiliated Hospital of North Sichuan Medical College.
Funding
No funding was received.
Disclosure
The authors declare that there are no conflicts of interest.
References
1. Wiesmueller F, Kopke J, Aust D, et al. Silenced ZNF154 is associated with longer survival in resectable pancreatic cancer. Int J Mol Sci. 2019;20(21):5437. doi:10.3390/ijms20215437
2. Hu Y, Qi MF, Xu QL, et al. Candidate tumor suppressor ZNF154 suppresses invasion and metastasis in NPC by inhibiting the EMT via Wnt/β-catenin signalling. Oncotarget. 2017;8:85749–85758. doi:10.18632/oncotarget.20479
3. Margolin G, Petrykowska H, Jameel N, Bell D, Young A, Elnitski L. Robust detection of DNA hypermethylation of ZNF154 as a pan-cancer locus with in silico modeling for blood-based diagnostic development. J Mol Diagn. 2016;18:283–298. doi:10.1016/j.jmoldx.2015.11.004
4. Reinert T, Borre M, Christiansen A, Hermann G, Ørntoft T, Dyrskjøt L. Diagnosis of bladder cancer recurrence based on urinary levels of EOMES, HOXA9, POU4F2, TWIST1, VIM, and ZNF154 hypermethylation. PLoS One. 2012;7(10):e46297. doi:10.1371/journal.pone.0046297
5. González CA, Agudo A. Carcinogenesis, prevention and early detection of gastric cancer: where we are and where we should go. Int J Cancer. 2012;130(4):745–753. doi:10.1002/ijc.26430
6. Cheung KF, Lam CN, Wu K, et al. Characterization of the gene structure, functional significance, and clinical application of RNF180, a novel gene in gastric cancer. Cancer. 2012;118:947–959. doi:10.1002/cncr.26189
7. Yu J, Cheng YY, Tao Q, et al. Methylation of protocadherin 10, a novel tumor suppressor, is associated with poor prognosis in patients with gastric cancer. Gastroenterology. 2009;136:640–651. doi:10.1053/j.gastro.2008.10.050
8. Germack R, Dickenson JM. Adenosine triggers preconditioning through MEK/ERK1/2 signalling pathway during hypoxia/reoxygenation in neonatal rat cardiomyocytes. J Mol Cell Cardiol. 2004;37:989–999. doi:10.1016/j.yjmcc.2004.08.001
9. Sung HJ, Kim Y, Kang H, et al. Inhibitory effect of trolox on the migration and invasion of human lung and cervical cancer cells. Int J Mol Med. 2012;29:245–251. doi:10.3892/ijmm.2011.832
10. Zhou B, Chen H, Wei D, et al. A novel miR-219-SMC4-JAK2/Stat3 regulatory pathway in human hepatocellular carcinoma. J Exp Clin Cancer Res. 2014;33(1):55. doi:10.1186/1756-9966-33-55
11. Das B, Chowdhury C, Kumar D, et al. Synthesis, cytotoxicity, and structure-activity relationship (SAR) studies of andrographolide analogues as anti-cancer agent. Bioorg Med Chem Lett. 2010;20:6947–6950. doi:10.1016/j.bmcl.2010.09.126
12. Smith DD, Schwarz RR, Schwartz RE. Impact of total lymph node count on staging and survival after gastrectomy for gastric cancer: data from a large US-population database. J Clin Oncol. 2005;23:7114–7124. doi:10.1200/JCO.2005.14.621
13. Lander ES, Linton LM, Birren B, et al. Initial sequencing and analysis of the human genome. Nature. 2001;409:860–921. doi:10.1038/35057062
14. Tupler R, Perini G, Green MR. Expressing the human genome. Nature. 2001;409(6822):832–833. doi:10.1038/35057011
15. Jen J, Wang YC. Zinc finger proteins in cancer progression. J Biomed Sci. 2016;23:53. doi:10.1186/s12929-016-0269-9
16. Ye Q, Liu J, Xie K. Zinc finger proteins and regulation of the hallmarks of cancer. Histol Histopathol. 2019;34:1097–1109. doi:10.14670/HH-18-121
17. Cheng Y, Liang P, Geng H, et al. A novel 19q13 nucleolar zinc finger protein suppresses tumor cell growth through inhibiting ribosome biogenesis and inducing apoptosis but is frequently silenced in multiple carcinomas. Mol Cancer Res. 2012;10:925–936. doi:10.1158/1541-7786.MCR-11-0594
18. Deng J, Liang H, Ying G, et al. Poor survival is associated with the methylated degree of zinc-finger protein 545 (ZNF545) DNA promoter in gastric cancer. Oncotarget. 2015;6:4482–4495. doi:10.18632/oncotarget.2916
19. Yu J, Liang QY, Wang J, et al. Zinc-finger protein 331, a novel putative tumor suppressor, suppresses growth and invasiveness of gastric cancer. Oncogene. 2013;32:307–317. doi:10.1038/onc.2012.54
20. Vedeld HM, Andresen K, Eilertsen IA, et al. The novel colorectal cancer biomarkers CDO1, ZSCAN18 and ZNF331 are frequently methylated across gastrointestinal cancers. Int J Cancer. 2015;136:844–853. doi:10.1002/ijc.29039
21. Jiang S, Linghu E, Zhan Q, Han W, Guo M. Methylation of ZNF331 promotes cell invasion and migration in human esophageal cancer. Curr Protein Pept Sci. 2015;16:322–328. doi:10.2174/138920371604150429155255
22. Hill VK, Ricketts C, Bieche I, et al. Genome-wide DNA methylation profiling of CpG islands in breast cancer identifies novel genes associated with tumorigenicity. Cancer Res. 2011;71:2988–2999. doi:10.1158/0008-5472.CAN-10-4026
23. Cancer Genome Atlas Research Network. Comprehensive genomic characterization of squamous cell lung cancers. Nature. 2012;489(7417):519–525. doi:10.1038/nature11404
24. Shen J, Wang S, Zhang YJ, et al. Genome-wide DNA methylation profiles in hepatocellular carcinoma. Hepatology. 2012;55:1799–1808. doi:10.1002/hep.25569
25. Kolbe DL, DeLoia JA, Porter-Gill P, et al. Differential analysis of ovarian and endometrial cancers identifies a methylator phenotype. PLoS One. 2012;7:e32941. doi:10.1371/journal.pone.0032941
26. Arai E, Chiku S, Mori T, et al. Single-CpG-resolution methylome analysis identifies clinicopathologically aggressive CpG island methylator phenotype clear cell renal cell carcinomas. Carcinogenesis. 2012;33:1487–1493. doi:10.1093/carcin/bgs177
27. Mahapatra S, Klee EW, Young CY, et al. Global methylation profiling for risk prediction of prostate cancer. Clin Cancer Res. 2012;18:2882–2895. doi:10.1158/1078-0432.CCR-11-2090
28. Sánchez-Vega F, Gotea V, Petrykowska HM, et al. Recurrent patterns of DNA methylation in the ZNF154, CASP8, and VHL promoters across a wide spectrum of human solid epithelial tumors and cancer cell lines. Epigenetics. 2013;8:1355–1372.
29. Carr I. Lymphatic metastasis. Cancer Metastasis Rev. 1983;2:307–317. doi:10.1007/BF00048483
30. Zhang X-Y, Lu W-Y. Recent advances in lymphatic targeted drug delivery system for tumor metastasis. Cancer Biol Med. 2014;11(4):247–254. doi:10.7497/j.issn.2095-3941.2014.04.003
 © 2022 The Author(s). This work is published and licensed by Dove Medical Press Limited. The full terms of this license are available at https://www.dovepress.com/terms.php and incorporate the Creative Commons Attribution - Non Commercial (unported, v3.0) License.
By accessing the work you hereby accept the Terms. Non-commercial uses of the work are permitted without any further permission from Dove Medical Press Limited, provided the work is properly attributed. For permission for commercial use of this work, please see paragraphs 4.2 and 5 of our Terms.
© 2022 The Author(s). This work is published and licensed by Dove Medical Press Limited. The full terms of this license are available at https://www.dovepress.com/terms.php and incorporate the Creative Commons Attribution - Non Commercial (unported, v3.0) License.
By accessing the work you hereby accept the Terms. Non-commercial uses of the work are permitted without any further permission from Dove Medical Press Limited, provided the work is properly attributed. For permission for commercial use of this work, please see paragraphs 4.2 and 5 of our Terms.