Back to Journals » Cancer Management and Research » Volume 10
Low expression of VSIG4 is associated with poor prognosis in hepatocellular carcinoma patients with hepatitis B infection
Authors Zhu S , Tan W, Li W, Zhou R, Wu X, Chen X, Li W, Shang C , Chen Y
Received 17 February 2018
Accepted for publication 28 May 2018
Published 20 September 2018 Volume 2018:10 Pages 3697—3705
DOI https://doi.org/10.2147/CMAR.S165822
Checked for plagiarism Yes
Review by Single anonymous peer review
Peer reviewer comments 2
Editor who approved publication: Professor Harikrishna Nakshatri
Sicong Zhu,1,2,* Wenliang Tan,1,2,* Wenxin Li,1,2 Rui Zhou,1,2 Xiaolin Wu,1,2 Xianqing Chen,1,2 Wenda Li,2 Changzhen Shang,2 Yajin Chen2
1Guangdong Provincial Key Laboratory of Malignant Tumor Epigenetics and Gene Regulation, Medical Research Center, Sun Yat-sen Memorial Hospital, Sun Yat-sen University, Guangzhou, People’s Republic of China; 2Department of Hepatobiliary Surgery, Sun Yat-sen Memorial Hospital, Sun Yat-sen University, Guangzhou, People’s Republic of China
*These authors contributed equally to this work
Background: V-set and immunoglobulin domain containing protein 4 (VSIG4) was reported to play an important role in tumorigenesis. However, the expression and clinical relevance in hepatocellular carcinoma (HCC) remain unknown.
Materials and methods: First, the mRNA profiles of HCC were screened from The Cancer Genome Atlas (TCGA) and Gene Expression Omnibus databases. VSIG4, a differentially expressed gene that has not been reported in HCC, was distinguished. Second, the correlation between VSIG4 expression and the prognosis of HCC patients from TCGA was analyzed. Third, VSIG4 mRNA level was detected in 36 pairs of HCC tissues and 4 HCC cell lines by PCR assay. And finally, prognosis analysis was assessed for 36 HCC patients with different expression levels of VSIG4.
Results: Bioinformatics analysis showed that VSIG4 expression was downregulated in HCC tissues, and the expression level of VSIG4 was negatively correlated with serum alpha fetal protein (AFP) level and tumor distant metastasis. Survival analysis of all HCC patients in TCGA indicated that the overall survival and disease-free survival were not significantly associated with VSIG4 expression. However, subgroup analysis showed that in the patients with hepatitis B virus-related HCC, both overall survival and disease-free survival were shorter in the low VSIG4 expression group. Our PCR results further showed that VSIG4 expression was significantly decreased in HCC tissues and HCC cell lines, and the disease-free survival in hepatitis B virus-related HCC patients with low VSIG4 expression was shorter than in those with high VSIG4 expression, which was consistent with the bioinformatics analysis results.
Conclusion: Our study suggests that VSIG4 is downregulated in HCC, and low expression of VSIG4 is associated with poor prognosis in hepatitis B virus-related HCC patients.
Keywords: hepatocellular carcinoma, VSIG4, hepatitis B infection, bioinformatics analysis
Introduction
Hepatocellular carcinoma (HCC) is the sixth most common cancer and the fourth main cause of cancer-related deaths worldwide.1 In recent years, great improvements have been made in the treatment of HCC, such as in liver transplantation, surgical resection, local ablation, and transcatheter arterial chemoembolization.2,3 However, the 5-year survival rate of HCC is still very low, mainly due to insidious onset of the disease and high rate of tumor recurrence as well as tumor metastasis.4 Therefore, a better understanding of HCC pathogenesis and prognosis prediction is urgently needed to refine the outcomes of HCC patients.
V-set and immunoglobulin domain containing protein 4 (VSIG4), also called CRIg and Z39Ig, is a novel type 1 transmembrane protein of B7-related immunoglobulin superfamily.5,6 The VSIG4 protein, which is mainly distributed in liver, heart, and placenta, is well known for the negative regulation effect of T-cells.7 VSIG4 was proved to be a coinhibitory ligand that negatively regulates T-cell activation through inhibiting CD4+ and CD8+ T-cell proliferation and IL-2 production.8 Recent studies have confirmed that VSIG4 gene was overexpressed on several kinds of cancer cells such as lung cancer, ovarian cancer, breast cancer, glioblastoma, and multiple myeloma, and acted as an oncogene that led to the functions of cellular immunity and promotion of tumor progression.9–13 In the research fields of liver diseases, relative studies have suggested that VSIG4 expressed on Kupffer cells could serve as a protective factor for liver injury.14 However, the expression and biological function of VSIG4 in HCC are still unknown.
The current study aimed to investigate the expression of VSIG4 in HCC and the prognostic significance of VSIG4 in patients with HCC. The bioinformatics analyses demonstrated that VSIG4 was downregulated in HCC tissues, and low expression level of VSIG4 was closely related to poor prognosis in hepatitis B virus (HBV)-positive HCC patients. However, VSIG4 expression has no markedly predictive significance in HBV-negative patients. Moreover, by exploring the VSIG4 expression in 4 HCC cell lines and 36 pairs of tissue specimens, we further confirmed the low expression of VSIG4 and its predictive significance for HCC, which were consistent with the bioinformatics analysis.
Materials and methods
Cell lines and cell culture
Four human hepatoma cell lines (HepG2, SK-hep-1, Huh-7, and MHCC-97H) were purchased from the Cell Bank of the Chinese Academy of Sciences (Shanghai, People’s Republic of China). The human liver cell line LO2 was obtained from our laboratory, and its use was approved by the ethics committee of Sun Yat-sen Memorial Hospital, Sun Yat-sen University (Guangzhou, People’s Republic of China). All cell lines were cultured in Dulbecco’s modified Eagle’s medium (Gibco, Grand Island, NY, USA) supplemented with 10% fetal bovine serum (Biological Industries, Haemek, Israel) and 100 units/mL penicillin plus 100 µg/mL streptomycin. All cells were maintained at 37°C in an atmosphere containing 5% CO2.
Patients and tumor tissues
HCC tissues and the adjacent normal tissues were collected from 36 patients during hepatectomy for HCC at Sun Yat-sen Memorial Hospital from 2016 to 2017. All patients were diagnosed based on the clinical and pathological evidences when being admitted to our center. The tissue specimens were immediately snap frozen in liquid nitrogen after resection from patients and then stored at –80°C. None of the patients received radiotherapy, chemotherapy, or transcatheter arterial chemoembolization or were treated with targeted-drugs such as sorafenib before surgery.
RNA extraction and quantitative reverse transcription polymerase chain reaction (qRT-PCR)
Total RNA from tissues or cell lines was extracted with Trizol reagent (Takara, Shiga, Japan) according to the manufacturer’s instructions. mRNA was reverse transcribed by using a PrimeScript RT reagent Kit (Takara). cDNA was amplified and quantified on CFX96 system (Bio-Rad, Hercules, CA, USA) using SYBR Premix Ex Taq (Takara). Briefly, PCRs were performed in a final volume of 10 µL, and the thermal conditions were 95°C for 30 seconds followed by 40 cycles of 95°C for 5 seconds and 60°C for 20 seconds. GAPDH was used as endogenous controls. Expression of VSIG4 was determined using the 2–ΔΔCT method.15 The primers were VSIG4 sense AACCACCATACAGCACAAGCA, antisense CAAGACCAAGACTGAGGCACC; GAPDH sense ACAACTTTGGTATCGTGGAAGG, antisense GCCATCACGCCACAGTTTC.
The Cancer Genome Atlas (TCGA) and Gene Expression Omnibus (GEO) data
TCGA database, which contains gene information through large-scale genomic sequencing, is a bank for multidimensional genomic information of 33 types of human cancers. Data including gene expression profiles and clinical information of HCC patients were obtained from the Genomic Data Commons Data Portal within TCGA data portal (https://portal.gdc.cancer.gov/). Of 427 TCGA mRNA data samples of liver cancer (377 tumors and 50 normals) downloaded from the database, those samples missing relevant clinical information were eliminated. When this was done, there were 232 samples (182 tumors, 50 normals) left, and those data were analyzed in our study. GEO is another genomics data repository of high-throughput gene expression data including hybridization arrays, chips, and microarrays. One series of gene expression profiling, including gene information of 60 patients with HCC, was downloaded to investigate aberrant gene expression in HCC.
Gene information acquisition and gene screening
The gene expression profiles of patients with HCC in TCGA were extracted from the database by R software (The R Foundation, Vienna, Austria), and gene expression levels of the samples (Fragments Per Kilobase of transcript per Million [FPKM] standardization) were obtained. Fifty patients, whose gene expression profiles included both tumor tissues and matched adjacent normal tissues, were chosen to the next step. The genes with differential expression in HCC were distinguished using the glmnet package of R software. Lasso (least absolute shrinkage and selection operator) regression analysis was performed following known procedures16,17 to further screen the genes to assess if the fold changes of expression between tumor and normal tissues were >2. Thus, those genes with a better research value for our study were acquired.
Statistical analysis
All of the statistical analysis including independent and paired t-test, rank sum test, lasso regression analysis, and survival analysis were done using SPSS 22.0 (IBM Corporation, Armonk, NY, USA) and R software. In our study, p-value <0.05 was considered significant.
Ethics approval
This study was approved by the ethics committee of Sun Yat-sen Memorial Hospital, Sun Yat-sen University. Informed consent was signed by all the patients whose tissue specimens were used in our study.
Results
Significant difference of VSIG4 expression in tumor and normal tissues
A number of genes were screened using the glmnet package of R software, based on p-value of paired t-test and Wilcoxon signed rank test (data not shown). After dimensionality reduction by lasso regression model, the top 30 mRNAs which were considered to have obvious statistical differences of expression in tumor and normal tissues were selected (Table S1). VSIG4, the gene that has not been studied in HCC, was one of these selected genes. Also, VSIG4 showed significant differential expression between tumor and normal tissues, and thus was worth further exploration.
The expression of VSIG4 in HCC, verified by bioinformatics analysis
The expression of VSIG4 mRNA was investigated by using the gene expression information of profiles in TCGA and GEO datasets. TCGA data showed that the expression of VSIG4 was significant lower in 50 HCC tissues, compared with that in 50 matched adjacent tissues (p<0.01) (Figure 1A). Similar results that VSIG4 expression was downregulated in tumors were obtained by analyzing GSE64041 dataset with 60 pairs of tissue samples (p<0.001) (Figure 1B).
Relevance between VSIG4 expression and clinicopathological characteristics
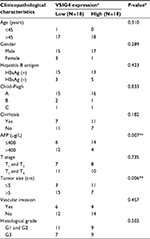
By analyzing the clinicopathological parameters of 182 HCC patients from the TCGA database, univariate analysis revealed that VSIG4 expression was significantly associated with serum level of AFP (p=0.022) and tumor M stage (p=0.009) (Table 1). But there was no obvious association with other characteristics including age, gender, viral hepatitis infection, Child–Pugh classification, cirrhosis, T and N stages, as well as tumor differentiation (p>0.05). Figure 1C and D shows that low expression level of VSIG4 was closely related to high serum AFP level (p=0.028) and tumor distant metastasis (p=0.029).
Correlation between VSIG4 expression and survival of HCC patients
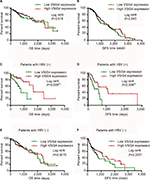
The survival curves for overall survival (OS) (Figure 2A) and disease-free survival (DFS) (Figure 2B) of 182 HCC patients in TCGA database were plotted. Although the median OS and DFS of patients with high VSIG4 expression was slightly longer than those with low VSIG4 expression, Kaplan–Meier analysis showed little relationship between VSIG4 expression and survival time (p all >0.05). Then, the patients were divided into several subgroups based on clinicopathological parameters including race, clinical stage, tumor histological grade, and viral hepatitis infection. We did not find significant differences between VSIG4 expression and survival time of patients when subdivided based on different race, clinical stage, and tumor histological grade (data not shown). But interestingly, in the HBV-positive HCC patients, we found that median OS in high VSIG4 expression group (1,694 days) was significantly longer than that in the low VSIG4 expression group (768 days) (Figure 2C). Also, it was demonstrated that patients with low VSIG4 mRNA level had a shorter DFS (Figure 2D). Univariate analysis also demonstrated that VSIG4 expression was associated with AFP level and tumor distant metastasis (Table S2). Cox analysis showed that VSIG4 expression was an independent prognostic factor in HBV-related HCC, for both OS and DFS (Table S3). However, Kaplan–Meier survival curves for OS (Figure 2E) and DFS (Figure 2F) still showed no significant differences in the HBV-negative HCC patients, including in patients with hepatitis C virus (HCV) infection (Figure S1A and B). We also found no significant correlation between VSIG4 expression and the clinicopathological parameters of HBV-negative HCC patients (Table S4), including those with HCV infection (Table S5).
VSIG4 expression in hepatoma cell lines and surgically removed HCC tissues
qRT-PCR assay was conducted to further confirm the phenomenon of VSIG4 downregulation in HCC. We compared the expression of VSIG4 in 4 human hepatoma cell lines including HepG2, Sk-hep-1, Huh7, and MHCC-97H, with that in human liver cell strain LO2. As can be seen from Figure 3D, VSIG4 expression in hepatoma cells was significantly lower than that in normal liver cells (all p<0.001). Then, we explored the mRNA level of VSIG4 in 36 pairs of HCC and adjacent tissues specimens which were surgically removed from 36 HCC patients in our center. The results of the PCR assay showed low expression of VSIG4 in 86.1% (31/36) HCC specimens, compared with matched adjacent specimens (Figure 3A). Univariate analysis of clinical characteristics of 36 patients indicated that VSIG4 expression was significantly correlated with tumor size and AFP level (Table 2). The expression level of VSIG4 was negatively related to the largest diameter of tumor (p=0.002) (Figure 3B) and serum level of AFP (p=0.011) (Figure 3C), which indicated a relationship between VSIG4 and prognosis of HCC patients.
Relationship between VSIG4 expression and prognosis of HCC patients
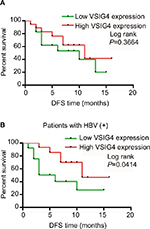
All the 36 patients with HCC were clinically followed up for >1 year. The DFS of the patients with low VSIG4 expression was not significantly different from that with high VSIG4 expression (Figure 4A). But in the HCC patients with HBV infection, the DFS of patients with low VSIG4 expression was significantly shorter than those with high VSIG4 expression (Figure 4B), which was consistent with the conclusion verified from TCGA data. HBV-positive HCC patients with low VSIG4 expression had poor prognosis, which suggested that VSIG4 was a potential tumor suppressor gene in HBV-related HCC.
Discussion
HCC is one of the most common and aggressive malignant tumors with poor clinical outcomes. The conventional clinical stage classification systems are helpful in predicting the prognosis of the patients with different pathologic types.18 However, due to the high heterogeneity of HCC, further research should be undertaken to seek novel factors that could predict the prognosis of HCC more effectively. In this study, we investigated the expression of VSIG4 in HCC, and assessed its function in predicting the outcome of HCC patients. We demonstrated that low VSIG4 expression was closely associated with poor prognosis of HCC patients, especially in those with HBV infection.
VSIG4 is a member of the B7 family-related protein which is well known for its inhibition of T-cell activation and works as a coinhibitory ligand that inhibits T-cell activation and cytokine production through cell cycle arrest at the G0/G1 phase and induces T-cell anergy.19 In previous research, it has been reported that VSIG4 can regulate the tumor microenvironment and promote the proliferation, invasion, and migration of breast cancer cells, which indicates that VSIG4 also makes a significant contribution to the development and progression of the tumor.10 Additionally, VSIG4 expression in ovarian cancer tissues was lower in advanced and recurrent ovarian cancer, which suggested that VSIG4 can weaken the malignancy of the tumor.11 Furthermore, VSIG4 acting as the immune checkpoint molecule can be used as an independent prognostic factor for multiple myeloma and high-grade glioma.12,13 However, the expression of VSIG4 and its association with clinicopathological characteristics in HCC remain unknown. In our study, we demonstrated that VSIG4 mRNA was downregulated in HCC by bioinformatics analysis, and low expression level of VSIG4 was closely related to high serum AFP level and tumor distance recurrence, which indicated poor prognosis of HCC patients. Moreover, we explored VSIG4 expression in 4 HCC cell lines and 36 pairs of tissue specimens in our center, and the results of the PCR assay were consistent with bioinformatics analysis. These results suggested that VSIG4 gene functioned as a tumor suppressor gene in HCC.
To date, the underlying mechanism of VSIG4 in HCC remains uncertain. It has been reported that VSIG4 knockout mice showed severe hepatitis in liver injury animal models.20 Thus, we speculated that there may exist a certain relationship between hepatitis and VSIG4 in the aspect of HCC pathogenesis.
Hepatitis B infection is also a reason for HCC. Previous studies have confirmed that the viral load of hepatitis B is a risk factor for HCC development in chronic hepatitis B (CHB) patients, and viral suppression could slow the progression of HCC.21 Another related study22 demonstrated the relation between TIP30 expression and HBV infection and proved that TIP30 was an independent poor prognostic factor for the OS of HCC patients. Our study found that OS and DFS were not significantly associated with VSIG4 expression in all HCC patients, based on the clinical data from TCGA. However, it is noteworthy that, in the subgroup of HBV-related HCC patients, the OS and DFS of the patients with low VSIG4 expression were shorter than in those with high VSIG4 expression. Cox regression analysis showed that VSIG4 expression was an independent prognostic factor for both OS and DFS. We also obtained the similar results in the HCC patients from our center that HBV-positive HCC patients with low VSIG4 expression had shorter DFS. Nevertheless, VSIG4 failed to predict the prognosis of the patients with HCV, the mechanism of hepatocarcinogenesis of which differs from HBV.23
Taken together, our present study demonstrated that the low expression of VSIG4 was closely related to poor prognosis in HBV-positive HCC patients. However, VSIG4 expression has no markedly predictive significance in HBV-negative patients. Therefore, VSIG4 gene might serve as a potential tumor suppressor gene and a novel biomarker for HBV-related HCC. As the exact underlying mechanism of VSIG4 in HCC remains uncertain, further studies concerning VSIG4 and HCC pathogenesis need to be performed.
Conclusion
To our knowledge, this study represents a first report identifying that VSIG4 was downregulated in HCC. Decreased expression of VSIG4 is an independent poor prognostic factor in HBV-related HCC. We suggest that VSIG4 could be a novel potential biomarker for HCC patients with HBV infection.
Acknowledgment
This work was supported by grants from the National Natural Science Foundation of China (nos. 81572398, 81672419), the Science and Technology Planning Project of Guangdong Province (nos. 2017A010105003, 2015A050502023, 2016A020216010), the Natural Science Foundation of Guangdong Province (nos. 2014A030313061, 2013B021800101), and the Techpool Research Foundation of China (no. UF201312).
Disclosure
The authors report no conflicts of interest in this work.
References
Yu WB, Rao A, Vu V, Xu L, Rao JY, Wu JX. Management of centrally located hepatocellular carcinoma: update 2016. World J Hepatol. 2017;9(13):627–634. | ||
Carr BI. Hepatocellular carcinoma: current management and future trends. Gastroenterology. 2004;127(5 Suppl 1):S218–S224. | ||
Llovet JM. Updated treatment approach to hepatocellular carcinoma. J Gastroenterol. 2005;40(3):225–235. | ||
Schoenberg MB, Bucher JN, Vater A, et al. Resection or transplant in early hepatocellular carcinoma. Dtsch Arztebl Int. 2017;114(31–32):519–526. | ||
Walker MG. Z39Ig is co-expressed with activated macrophage genes. Biochim Biophys Acta. 2002;1574(3):387–390. | ||
Ahn JH, Lee Y, Jeon C, et al. Identification of the genes differentially expressed in human dendritic cell subsets by cDNA subtraction and microarray analysis. Blood. 2002;100(5):1742–1754. | ||
Helmy KY, Katschke KJ, Gorgani NN, et al. CRIg: a macrophage complement receptor required for phagocytosis of circulating pathogens. Cell. 2006;124(5):915–927. | ||
Vogt L, Schmitz N, Kurrer MO, et al. VSIG4, a B7 family-related protein, is a negative regulator of T cell activation. J Clin Invest. 2006;116(10):2817–2826. | ||
Liao Y, Guo S, Chen Y, et al. VSIG4 expression on macrophages facilitates lung cancer development. Lab Invest. 2014;94(7):706–715. | ||
Sturtz LA, Deyarmin B, van Laar R, et al. Gene expression differences in adipose tissue associated with breast tumorigenesis. Adipocyte. 2014;3(2):107–114. | ||
Byun JM, Jeong DH, Choi IH, et al. The significance of VSIG4 expression in ovarian cancer. Int J Gynecol Cancer. 2017;27(5):872–878. | ||
Xu T, Jiang Y, Yan Y, et al. VSIG4 is highly expressed and correlated with poor prognosis of high-grade glioma patients. Am J Transl Res. 2015;7(6):1172–1180. | ||
Roh J, Jeon Y, Lee AN, et al. The immune checkpoint molecule V-set Ig domain-containing 4 is an independent prognostic factor for multiple myeloma. Oncotarget. 2017;8(35):58122–58132. | ||
Jung K, Kang M, Park C, et al. Protective role of V-set and immunoglobulin domain-containing 4 expressed on Kupffer cells during immune-mediated liver injury by inducing tolerance of liver T- and natural killer T-cells. Hepatology. 2012;56(5):1838–1848. | ||
Livak KJ, Schmittgen TD. Analysis of relative gene expression data using real-time quantitative PCR and the 2–ΔΔCT method. Methods. 2001;25(4):402–408. | ||
Tibshirani R. The lasso method for variable selection in the Cox mode. Stat Med. 1997;16(4):385–395. | ||
Segal MR, Dahlquist KD, Conklin BR. Regression approaches for microarray data analysis. J Comput Biol. 2003;10(6):961–980. | ||
Ding XX, Zhu QG, Zhang SM, et al. Precision medicine for hepatocellular carcinoma: driver mutations and targeted therapy. Oncotarget. 2017;8(33):55715–55730. | ||
Vogt L, Schmitz N, Kurrer MO, et al. VSIG4, a B7 family-related protein, is a negative regulator of T cell activation. J Clin Invest. 2006;116(10):2817–2826. | ||
Jung K, Kang M, Park C, et al. Protective role of V-set and immunoglobulin domain-containing 4 expressed on Kupffer cells during immune-mediated liver injury by inducing tolerance of liver T- and natural killer T-cells. Hepatology. 2012;56(5):1838–1848. | ||
Zhang W, Wang X, Wang Y, et al. Effective viral suppression is necessary to reduce hepatocellular carcinoma development in cirrhotic patients with chronic hepatitis B: results of a 10-year follow up. Medicine (Baltimore). 2017;96(44):e8454. | ||
Zhang X, Lv L, Ouyang X, et al. Association of TIP30 expression and prognosis of hepatocellular carcinoma in patients with HBV infection. Cancer Med. 2016;5(9):2180–2189. | ||
He B, Zhang H, Shi T. A comprehensive analysis of the dynamic biological networks in HCV induced hepatocarcinogenesis. PLoS One. 2011;6(4):e18516. |
 © 2018 The Author(s). This work is published and licensed by Dove Medical Press Limited. The full terms of this license are available at https://www.dovepress.com/terms.php and incorporate the Creative Commons Attribution - Non Commercial (unported, v3.0) License.
By accessing the work you hereby accept the Terms. Non-commercial uses of the work are permitted without any further permission from Dove Medical Press Limited, provided the work is properly attributed. For permission for commercial use of this work, please see paragraphs 4.2 and 5 of our Terms.
© 2018 The Author(s). This work is published and licensed by Dove Medical Press Limited. The full terms of this license are available at https://www.dovepress.com/terms.php and incorporate the Creative Commons Attribution - Non Commercial (unported, v3.0) License.
By accessing the work you hereby accept the Terms. Non-commercial uses of the work are permitted without any further permission from Dove Medical Press Limited, provided the work is properly attributed. For permission for commercial use of this work, please see paragraphs 4.2 and 5 of our Terms.