Back to Journals » International Journal of General Medicine » Volume 14
Low Expression of PLAT in Breast Cancer Infers Poor Prognosis and High Immune Infiltrating Level
Authors Wang X, Xue D, Zhu X, Geng R, Bao X, Chen X, Xia T
Received 26 October 2021
Accepted for publication 8 December 2021
Published 22 December 2021 Volume 2021:14 Pages 10213—10224
DOI https://doi.org/10.2147/IJGM.S341959
Checked for plagiarism Yes
Review by Single anonymous peer review
Peer reviewer comments 2
Editor who approved publication: Dr Scott Fraser
Xinyang Wang,1,* Dandan Xue,1,* Xiaoxia Zhu,2,* Rui Geng,3 Xin Bao,2 Xiang Chen,2 Tiansong Xia1,4
1Department of Breast Surgery, The First Affiliated Hospital with Nanjing Medical University, Nanjing, People’s Republic of China; 2Department of Thyroid and Breast, Yixing People’s Hospital Affiliated to Jiangsu University, Yixing, People’s Republic of China; 3Department of Biostatistics, School of Public Health, Nanjing Medical University, Nanjing, People’s Republic of China; 4Jiangsu Key Lab of Cancer Biomarkers, Prevention, and Treatment, Jiangsu Collaborative Innovation Center for Cancer Personalized Medicine, School of Public Health, Nanjing Medical University, Nanjing, People’s Republic of China
*These authors contributed equally to this work
Correspondence: Xiang Chen
Department of Thyroid and Breast, Yixing People’s Hospital Affiliated to Jiangsu University, 75 Zhen’guan Road, Yixing, 214200, People’s Republic of China
Email [email protected]
Tiansong Xia
Department of Breast Surgery, The First Affiliated Hospital with Nanjing Medical University, 300 Guangzhou Road, Nanjing, 210029, People’s Republic of China
Email [email protected]
Purpose: Breast cancer accounts for the highest incidence of tumors in women. Immune infiltrating of the tumor microenvironment positively correlates with the overall survival of breast cancer patients. PLAT can affect the development of many cancers, but its mechanism in breast cancer is unclear. We assessed the correlation between PLAT and immune infiltrating in breast cancer based on the TCGA database.
Patients and Methods: The expression and DNA methylation of PLAT in breast cancer with different clinical characteristics was tested by Wilcoxon signed rank test and displayed by box plot. Sequentially, Kaplan–Meier plot was employed to compare the difference in overall survival rates between patients with different expressed levels. Univariate and multivariate Cox regression analyses were used to validate whether PLAT is an independent prognostic factor of breast cancer. After that, GO, KEGG, and gene-set enrichment analysis were employed to do functional enrichment analysis. Finally, TIMER, TISIDB database, and ssGSEA algorithm were used to assess the correlation between PLAT expression and various immune characteristics. The correlation between PLAT expression and DNA methylation was examined by Pearson correlation coefficient.
Results: PLAT displays differential expression levels in breast cancer patients with various clinical characteristics. As an independent protective factor for breast cancer, PLAT may significantly correlate with the immune status of breast cancer by adjusting many immune molecules and affecting the immune infiltration in the tumor microenvironment. DNA methylation of PLAT downregulates the gene expression and affects the prognosis of breast cancer.
Conclusion: PLAT can be considered a potential biomarker to predict breast cancer prognosis and might contribute to the development of immunological treatment strategies.
Keywords: tPA, prognosis, immune infiltration, bioinformatic
Introduction
Breast cancer (BC) accounts for the highest incidence of tumors globally and is the leading cause of death of female cancer patients.1 In recent years, the application of precise surgery and adjuvant systemic treatments (chemotherapy, radiotherapy, endocrine therapy, and molecular targeting drugs) dramatically enhances the therapeutic effect. The prognosis of BC patients remains unsatisfactory, however. Screening and diagnosis are essential to reduce BC patients’ incidence rate and mortality rate in the early stage.2 Although clinical, pathological, and molecular features are commonly used to predict prognosis, the underlying pathogenesis of BC invasiveness is still poorly understood. Minimally invasive biomarkers to detect early BC and gauge treatment response are crucial in BC research.3
Tumor microenvironment (TME) plays a vital role in tumor progression and the overall efficacy of cancer treatments, such as immunotherapy, chemotherapy, and radiotherapy.4,5 The abundance of immune infiltrating cells in TME affects tumor initiation, progression, metastasis, and treatment resistance.6
Tissue type plasminogen activator (PLAT) and urokinase type plasminogen activator (PLAU), two essential proteins of the plasmin–plasminogen system, not only participated in the intravascular dissolution of blood clots but also played a significant role in tumor progression and metastasis.7 PLAU has been involved in different steps of cancer progression, such as tumorigenesis,8 angiogenesis,9 cell invasion, and metastasis.10 PLAT can affect the development of many tumors. BC patients with low PLAT levels often have a poor prognosis, while PLAT levels are higher in melanoma, neuroblastoma, acute myeloid leukemia, and pancreatic cancer.11,12
Our study evaluated the expression level of PLAT in BC and the relationships between PLAT expression and clinical pathological characteristics, together with prognosis, through publicly available data from The Cancer Genome Atlas (TCGA). PLAT protein expression was assessed with UALCAN databases. Moreover, the possible molecular functions of PLAT were screened by using GSEA software (version 4.1.0). Furthermore, the relationships between PLAT and immune infiltrating cells were explored using TIMER, TISIDB database, and ssGSEA algorithm. We investigated the correlation between PLAT expression level, clinicopathological characteristics, and DNA methylation with TCGA data. Our findings reveal the predictive value of PLAT in BC and show a potential correlation between PLAT and immune infiltrating cells, which will help clarify a possible mechanism for BC.
Materials and Methods
Data Acquisition
Transcriptional data of mRNA-sequencing, DNA methylation data, phenotype, and survival profiles of BC patients were accessed from UCSC Xena (https://xenabrowser.net/),13 which analyzes data from TCGA. The pre-processing steps were conducted by R (version 4.1.0) and Perl (version 5.30.2) software.
Gene Expression and DNA Methylation Analysis
We selected 1071 primary BC samples, and 96 paired normal samples with clinicopathological profiles such as age, sex, menopausal status, histological type, TNM stage, and PAM50 subtype14 for further analysis. PAM50 algorithm using the 50-gene classifier divides BC into five “intrinsic” subtypes: luminal A, luminal B, HER2-enriched, basal-like, and normal-like. We explored the different expressions of PLAT in all clinicopathologic features. PLAT methylation data of 890 samples and its correlated gene expression levels were applied to calculate the methylation score of PLAT and the relationships between gene expression, DNA methylation, and clinicopathologic profiles.
Survival Analysis
Profiles of overall survival (OS) and progression-free survival (PFS) were downloaded from TCGA survival data. From this data, 1071 samples were assigned to high and low expression groups based on the median score of PLAT. R “survival” package was employed to generate Kaplan–Meier (K–M) curves, while the optimal cut-off point for survival curves was generated through the “res. cut” function in the “survminer” package. Univariable and multivariable Cox regression models were established through the coxph function in the “survival” package, which was used to confirm whether PLAT is an independent prognostic factor of BC.
DEGs and Functional Enrichment Analysis
The “clusterProfiler” R package15 was employed to perform gene ontology (GO) and Kyoto Encyclopedia of Genes and Genomes (KEGG) analyses on the basis of differentially expressed genes (DEGs_ (| log2FC | ≥ 1, FDR < 0.05) between two different expressed groups. P-values were adjusted by the BH method. We used Spearman correlation coefficient analysis to show the relationship between PLAT and co-expression genes. GSEA was conducted based on KEGG (v7.4) gene-set collections using GSEA software (v4.1.0 6)16,17 to uncover pathways in the DEGs groups. Gene sets with NOM p-value < 0.05 were considered significant.
Characterization of the Tumor Microenvironment
The “gsva” package18 was utilized to conduct the ssGSEA,19 which can be applied to evaluate the scores of immune infiltrating cells and calculate the activity of immune-related pathways. TIMER (https://cistrome.shinyapps.io/timer/)20 contains an abundant resource for systematically exploring the immune infiltrates between various tumors. We employed TIMER 2.0 “gene” module to calculate the relationships between PLAT mRNA expression and the infiltration of immune cells. TISIDB database (http://cis.hku.hk/TISIDB)21 is a portal web containing a variety of tumor and immune system resources. In this study, TISIDB providers the correlations between PLAT and the immune system.
Statistical Analysis
Gene expression profiles, methylation degree, and clinical characteristics were compared through Wilcoxon signed rank tests and displayed by box plot. Survival rates were assessed by applying K–M curves and the log rank test. The Pearson chi-square test was employed to analyze the correlation between PLAT expression and DNA methylation level. R version 4.1.0 was used in all tests conducted. A statistically significant difference was considered when p < 0.05.
Results
Differential Expression of PLAT
We achieved PLAT expression and BC-related clinical profiles from the TCGA database. The detailed data are summarized in Supplementary Table 1. PLAT had a lower expression in BC tissues than in normal tissues both in TCGA-BRCA all samples (Figure 1A) and paired samples (Figure 1B). The CPTAC dataset of the UALCAN database was used to analyze the PLAT protein expression. PLAT in BC tissues was significantly lower than in normal tissues, which was consistent with the gene expression level (Figure 1C). After that, PLAT expression in BC with different clinical characteristics such as gender, menopausal status, histological type, molecular subtype, N stage, and T stage displayed statistical differences. The expression level of PLAT in women was lower than that in men (Figure 1D), and it was lower in postmenopausal women than premenopausal women (Figure 1E). Invasive ductal carcinoma (IDC) and invasive lobular carcinoma (ILC) are the two main histological types of BC. The expression level of PLAT in IDC was lower than in other types, including ILC. But there was no significant difference between ILC and other types (Figure 1F). Among the five PAM50 molecular subtypes of BC, luminal A, luminal B, HER2-enriched, basal-like, and normal-like, the expression level of PLAT was statistically different, in which luminal A was the highest and basal-like was the lowest (Figure 1G). The expression level was lower in the advanced stage in the T stage (Figure 1H); however, it was higher in the advanced N stage (Figure 1I).
PLAT Was an Independent Favorable Prognostic Factor
We explored whether PLAT expression was correlated with the prognosis of BC patients. K–M curves were utilized to evaluate the influence of PLAT expression on OS and DFS. The high PLAT expression group has a longer OS than low expression patients (Figure 2A) but there was no statistical significance in DFS (Figure 2B). We further investigated OS in the different molecular subtypes. Statistical significance of OS difference was found in all luminal samples, especially in luminal B subtypes (Figure 2C).
Univariate and multivariate regression analyses were applied to evaluate whether PLAT is an independent prognostic factor for BC. Univariate Cox hazard analysis identified that age (HR = 1.033, 95% CI:1.020–1.046, p < 0.001), menopausal status (HR = 2.025, 95% CI:1.347–3.045, p < 0.001), T stage (HR = 1.459, 95% CI:1.197–1.779, p < 0.001), N stage (HR = 1.600, 95% CI:1.347–1.901, p <0.001), M stage (HR = 4.860, 95% CI:2.900–8.144, p < 0.001), Stage (HR = 2.187, 95% CI:1.744–2.742, p < 0.001), and PLAT (HR = 0.886, 95% CI:0.797–0.984, p = 0.024) were prognostic factors of BRCA. Multivariate analysis revealed that, after excluding other confounding factors, age (HR = 1.044, 95% CI:1.023–1.067, p < 0.001), N stage (HR = 1.451, 95% CI:1.052–2.000, p = 0.023), and PLAT (HR = 0.850, 95% CI:0.742–0.974, p = 0.020) were still prognostic factors of BCRA (Figure 2D). The above results show that PLAT is an independent prognostic factor for BC and is a protective factor.
Enrichment Analysis
All TCGA–BRCA samples were assigned to two groups in the light of PLAT expression to gain insight into PLAT biological clues in BC. The volcano plot and heatmap show DEGs between the two groups (Figure 3A and B). PLAT and its co-expression genes are shown in Figure 3C. GO, and KEGG enrichment analyses were conducted based on 93 genes that had a positive correlation with PLAT to identify PLAT-related pathways and biological functions. The top 30 enriched GO terms are shown in Figure 3D. We can see that PLAT was mainly enriched in the pathway related to response to steroid hormone, humoral immune response, intracellular steroid hormone receptor signaling pathway, steroid hormone mediated signaling pathway, and response to progesterone. KEGG enrichment showed that PLAT was closely related to the pathways that correlate with BC and the immune response, such as the estrogen signaling pathway, apelin signaling pathway, IL-17 signaling pathway, and BC (Figure 3E).
We further used GSEA to predict PLAT-related pathways between patients with different PLAT expressions; 70 of 178 gene sets were up-regulated in the low PLAT group, and 15 pathways were enriched at NOM p < 0.05 and FDR < 0.25. The presenting figures include antigen processing and presentation, natural killer cell mediated cytotoxicity, homologous recombination, and nucleotide excision repair (Figure 3F). These results show that PLAT may affect immune cell infiltration.
Analysis of Immune Characteristics
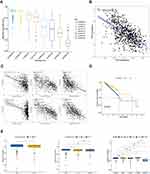
Immune characteristics are closely related to tumor progression. Through the functional analyses, we evaluated the enrichment scores of 16 immune cells and the activity of 13 immune-related pathways between the two expressed groups by employing ssGSEA. The low expression subgroup of PLAT generally had higher levels of infiltration of immune cells, especially of aDCs, CD8 + T cells, DCs, iDCs, macrophages, pDCs, Tfh, Th1 cells, Th2 cells, TILs, and Treg cells. Except for the parainflammation pathway, the other 12 immune pathways performed higher activity in the low expression group than in the high expression group (Figure 4A). After that, we evaluated the relationship between the PLAT expression in BRCA and immune cell infiltration in TIMER. The finding revealed that the expression level of PLAT had a negative correlation with the infiltration of neutrophil (Rho = −0.175, p = 2.59e-08), T cell cd4+ (Rho = −0.158, p = 5.16e-07), B cell (Rho = −0.117, p = 2.12e-04) and dendritic cell (Rho = −0.098, p = 2.08e-03). Meanwhile it had a positive correlation with the infiltration level of T cell CD8+ (Rho = 0.2, p = 2.13e-10) and macrophage (Rho = 0.266, p = 1.45e-17). Not surprisingly, tumor purity was negatively correlated with PLAT expression level (Figure 4B). Then, to understand the association between PLAT and immune infiltration, TISIDB was employed to evaluate the association between PLAT expression and various immune characteristics. Figure 4C shows the relationship between PLAT and tumor-infiltrating lymphocytes (TILs) like CD4, CD8, DC, and B cell and Figure 4D the relationship between PLAT and immunostimulators like IL2RA, ICOS, TNFRSF13C, and PVR. Figure 4E shows the relationship between PLAT and immunoinhibitors like CD274, PDCD1, PDCD1LG2, and CTLA4 and Figure 4F the relationships between PLAT and MHC molecules like TAP2, HLA-DOB, TAP1, and HLA-F. Figure 4G shows the relationship between PLAT and chemokines like CXCL10, CCL8, CCL18, and CCL7. Figure 4H shows the relationship between PLAT and chemokine receptors like CCR8, CXCR6, CCR1, and CCR5. The results mentioned above indicate that PLAT widely participates in adjusting many immune molecules in BC and influences the immune infiltration in TME.
Methylation Had a Negative Correlation with Gene Expression
After the studies mentioned above, we continued to explore the possible reasons for the difference in PLAT expression. The box plot shows the methylation degree of each methylation site of PLAT DNA. Sites cg03960326 and cg18460120 had a higher degree of methylation and sites cg22038738 and cg04563438 had lower methylation degree (Figure 5A). Subsequently, the association between DNA methylation level and gene expression was calculated. Figure 5B shows that there is a negative association between DNA methylation and gene expression on the whole. When analyzing a single site, it was found that the methylation levels of cg13880167, cg12091331, cg04563438, cg03960326, cg00491021, and cg22038738 were negatively correlated with gene expression (Figure 5C). After that, survival analysis was further conducted according to the methylation degree of each site. It was found that, among the six DNA methylation sites of PLAT that had a negative correlation with gene expression, the methylation degree of the cg03960326 site was significantly related to OS (Figure 5D). We further assessed the methylation differences of the cg03960326 site of PLAT DNA in different clinical feature groups. We can see that there are no distinctions in methylation levels between different TNM and stages. However, there existed differences in methylation levels of cg03960326 site in people with different menopause status, tumor histological types, and PAM50 molecular types (Figure 5E). These differences are contrary to the previous differences of PLAT gene in cases with different clinical features, which also verifies the negative relationship between DNA methylation and gene expression.
Discussion
PLAT, a serine protease, mainly takes part in the intravascular dissolution of blood clots.22 Previous studies have shown that PLAT promoter hypermethylation can down-regulate the expression of nasal polyps gene, resulting in excessive fibrin deposition, which can be used as a therapeutic target of NP.23 PLAT can also promote angiogenesis by mobilizing CD11 + cells.24 It can be applied to the treatment of acute stroke25,26 and non-small cell lung cancer.27 It is also an estrogen-induced protein in the human BC cell line.11 BC is one of the highest incidence rate diseases in the world, which has a poor prognosis. Early detection and timely treatment are the most effective methods to improve the survival rate of BC patients. Traditional prognostic factors include tumor type, grade, size, and lymph node status. They can provide some course information, but they are not very accurate. Tumor immunotherapy provides a new remedy option for BC. Therefore, we assessed the role of PLAT in BRCA.
We analyzed BC cases in the context of RNA-seq data, methylation profiles, and clinical characteristics achieved from TCGA. The findings revealed that the expression level of PLAT was lower in BC patients. And patients in an advanced stage or with a more aggressive histological type may have lower expression levels in general. Consistent with that, lower expression of PLAT in TCGA–BRCA profiles was related to shorter overall survival, especially in the luminal B subtype. So, we deem that higher PLAT expression is related to a better prognosis of BC. After that, univariate and multivariate Cox regression analyses showed that PLAT was an independent prognostic factor affecting the prognosis of BC patients and could be used as a predictive marker.
The enrichment analysis showed that PLAT was related to the immune and tumor-associated pathway. Infiltrating immune cells are an essential component of TME.28 Recently, immunotherapy for the interaction between immune cells and BC cells has been developed as an alternative to classical anticancer therapy.29,30 PLAT has been affirmed to predict the prognosis of multiple tumor types such as colon cancer, ovarian cancer, lung cancer, and BC.31,32 There is evidence proving that tumor-infiltrating lymphocytes (TIL) that existed before the treatment of BC can predict treatment response and improve prognosis.33 TIL plays a vital role in chemotherapy response and clinical prognosis of all subtypes of BC.34 In our study, the low expression subgroup of PLAT generally had higher levels of infiltration of immune cells and higher activity of immune-related pathways. PLAT expressed level had a negative correlation with the infiltration level of neutrophil, T cell CD4+, dendritic cell, and B cell. Meanwhile, it had a positive correlation with the infiltration level of T cell CD8+ and macrophage. PLAT expression is closely related to TILs, immunomodulatory factors, and chemokines. Our results verified the association between PLAT and the prognosis and immune infiltration of BC, suggesting that PLAT can help predict treatment response and improve prognosis.
DNA methylation plays a vital role in gene-expressed regulation, mainly related to transcriptional inhibition.35 Therefore, we continue to search for the possible causes of different gene expressions. Our results showed a negative relationship between the degree of DNA methylation and gene expression level. By further analyzing the methylation level of PLAT DNA in the cg03960326 site, which is significantly related to OS, methylation differences were observed in patients with different tumor histological types, molecular subtypes, and menopause. These differences are opposite to the previous differences of the PLAT gene in other clinical feature groups. This also verified that DNA methylation was negatively related to PLAT expression, which might cause PLAT expression differences.
Immune infiltration in the TME has been shown to play a critical role in cancer progression and occurrence and to affect clinical outcomes of cancer patients.36 In triple-negative BC, tumor-associated macrophages derived from peripheral blood monocytes promote tumor growth and progression by several mechanisms that include the secretion of inhibitory cytokines, the reduction of effector functions of TIL, and the promotion of Treg cells. Besides, tumor-associated macrophages have been shown to directly and indirectly modulate PD-1/PD-L1 expression in tumor environment.37 Immunotherapy has raised significant concern in the treatment of cancer, which aims to eliminate cancer cells by enhancing natural defenses. The blockade of PD-1/ PD-L1 and CTLA4 has achieved remarkable therapeutic success in multiple kinds of cancer, including BC. An in-depth understanding of immune infiltration cells in the TME is essential to uncover the underlying molecular mechanisms and provide new strategies to improve immunotherapeutic efficacy.
Previous analysis showed that PLAT, a tumor suppressor gene, has differential expression levels and independent prognostic value and is significantly correlated with tumor-related pathways and immune infiltrating cells in TME. However, one study has shown that PLAT cannot foresee the response to systemic adjuvant therapy.11 The presented research only shows the results of online databases. To further confirm this conclusion, we will conduct PCR and IHC on our clinical samples and further analyze the different clinical characteristics and survival information. Finally, the internal molecular mechanism of PLAT and tumor immune cell infiltration will be further explored. Therefore, further research is needed to improve the proof of the biological impact of PLAT.
Conclusion
PLAT can be considered a prognostic factor and biomarker to provide personalized treatment, improve the survival rate, and contribute to developing immunological treatment strategies.
Abbreviations
BC, breast cancer; TME, tumor microenvironment; PLAT, tissue-type plasminogen activator; PLAU, urokinase type plasminogen activator; TCGA, The Cancer Genome Atlas; OS, overall survival; PFS, progression-free survival; GO, Gene Ontology; KEGG, Kyoto Encyclopedia of Genes and Genomes; IDC, invasive ductal carcinoma; ILC, invasive lobular carcinoma; TIL, tumor-infiltrating lymphocytes.
Ethics Approval
This study was approved by the institutional ethical committee of the First Affiliated Hospital with Nanjing Medical University.
Acknowledgments
We would like to acknowledge the TCGA, TIMER, and TISIDB databases for free use.
Funding
This study is supported by the Natural Science Foundation of Jiangsu Province (BK20171506).
Disclosure
The authors report no conflicts of interest in this work.
References
1. Sung H, Ferlay J, Siegel RL, et al. Global cancer statistics 2020: GLOBOCAN estimates of incidence and mortality worldwide for 36 cancers in 185 countries. CA Cancer J Clin. 2021;71(3):209–249. doi:10.3322/caac.21660
2. Mayor S. Screening for early breast cancer reduces invasive cancer, study finds. BMJ. 2015;351:h6576. doi:10.1136/bmj.h6576
3. Arnedos M, Roulleaux Dugage M, Perez-Garcia J, Cortes J. Window of opportunity trials for biomarker discovery in breast cancer. Curr Opin Oncol. 2019;31(6):486–492. doi:10.1097/cco.0000000000000583
4. Galluzzi L, Buqué A, Kepp O, Zitvogel L, Kroemer G. Immunological effects of conventional chemotherapy and targeted anticancer agents. Cancer Cell. 2015;28(6):690–714. doi:10.1016/j.ccell.2015.10.012
5. Junttila MR, de Sauvage FJ. Influence of tumour micro-environment heterogeneity on therapeutic response. Nature. 2013;501(7467):346–354. doi:10.1038/nature12626
6. Pitt JM, Marabelle A, Eggermont A, Soria JC, Kroemer G, Zitvogel L. Targeting the tumor microenvironment: removing obstruction to anticancer immune responses and immunotherapy. Ann Oncol. 2016;27(8):1482–1492. doi:10.1093/annonc/mdw168
7. Mahmood N, Mihalcioiu C, Rabbani SA. Multifaceted role of the urokinase-type plasminogen activator (uPA) and its receptor (uPAR): diagnostic, prognostic, and therapeutic applications. Front Oncol. 2018;8:24. doi:10.3389/fonc.2018.00024
8. Duffy MJ. The urokinase plasminogen activator system: role in malignancy. Curr Pharm Des. 2004;10(1):39–49. doi:10.2174/1381612043453559
9. Su SC, Lin CW, Yang WE, Fan WL, Yang SF. The urokinase-type plasminogen activator (uPA) system as a biomarker and therapeutic target in human malignancies. Expert Opin Ther Targets. 2016;20(5):551–566. doi:10.1517/14728222.2016.1113260
10. Lester RD, Jo M, Montel V, Takimoto S, Gonias SL. uPAR induces epithelial-mesenchymal transition in hypoxic breast cancer cells. J Cell Biol. 2007;178(3):425–436. doi:10.1083/jcb.200701092
11. Corte MD, Vérez P, Rodríguez JC, et al. Tissue-type plasminogen activator (tPA) in breast cancer: relationship with clinicopathological parameters and prognostic significance. Breast Cancer Res Treat. 2005;90(1):33–40. doi:10.1007/s10549-004-2624-x
12. Díaz VM, Planaguma J, Thomson TM, Reventós J, Paciucci R. Tissue plasminogen activator is required for the growth, invasion, and angiogenesis of pancreatic tumor cells. Gastroenterology. 2002;122(3):806–819. doi:10.1053/gast.2002.31885
13. Cline MS, Craft B, Swatloski T, et al. Exploring TCGA pan-cancer data at the UCSC cancer genomics browser. Sci Rep. 2013;3:2652. doi:10.1038/srep02652
14. Parker JS, Mullins M, Cheang MC, et al. Supervised risk predictor of breast cancer based on intrinsic subtypes. J Clin Oncol. 2009;27(8):1160–1167. doi:10.1200/JCO.2008.18.1370
15. Yu G, Wang LG, Han Y, He QY. clusterProfiler: an R package for comparing biological themes among gene clusters. OMICS. 2012;16(5):284–287. doi:10.1089/omi.2011.0118
16. Mootha VK, Lindgren CM, Eriksson KF, et al. PGC-1α-responsive genes involved in oxidative phosphorylation are coordinately downregulated in human diabetes. Nat Genet. 2003;34(3):267–273. doi:10.1038/ng1180
17. Subramanian A, Tamayo P, Mootha VK, et al. Gene set enrichment analysis: a knowledge-based approach for interpreting genome-wide expression profiles. Proc Natl Acad Sci U S A. 2005;102(43):15545–15550. doi:10.1073/pnas.0506580102
18. Hänzelmann S, Castelo R, Guinney J. GSVA: gene set variation analysis for microarray and RNA-seq data. BMC Bioinform. 2013;14:7. doi:10.1186/1471-2105-14-7
19. Barbie DA, Tamayo P, Boehm JS, et al. Systematic RNA interference reveals that oncogenic KRAS-driven cancers require TBK1. Nature. 2009;462(7269):108–112. doi:10.1038/nature08460
20. Li T, Fu J, Zeng Z, et al. TIMER2.0 for analysis of tumor-infiltrating immune cells. Nucleic Acids Res. 2020;48(W1):W509–w514. doi:10.1093/nar/gkaa407
21. Ru B, Wong CN, Tong Y, et al. TISIDB: an integrated repository portal for tumor-immune system interactions. Bioinformatics. 2019;35(20):4200–4202. doi:10.1093/bioinformatics/btz210
22. Kruithof EK, Dunoyer-Geindre S. Human tissue-type plasminogen activator. Thromb Haemost. 2014;112(2):243–254. doi:10.1160/th13-06-0517
23. Kidoguchi M, Noguchi E, Nakamura T, et al. DNA methylation of proximal PLAT promoter in chronic rhinosinusitis with nasal polyps. Am J Rhinol Allergy. 2018;32(5):374–379. doi:10.1177/1945892418782236
24. Ohki M, Ohki Y, Ishihara M, et al. Tissue type plasminogen activator regulates myeloid-cell dependent neoangiogenesis during tissue regeneration. Blood. 2010;115(21):4302–4312. doi:10.1182/blood-2009-08-236851
25. Peña-Martínez C, Durán-Laforet V, García-Culebras A, et al. Pharmacological modulation of neutrophil extracellular traps reverses thrombotic stroke tPA (tissue-type plasminogen activator) resistance. Stroke. 2019;50(11):3228–3237. doi:10.1161/strokeaha.119.026848
26. Zhu J, Wan Y, Xu H, Wu Y, Hu B, Jin H. The role of endogenous tissue-type plasminogen activator in neuronal survival after ischemic stroke: friend or foe? Cell Mol Life Sci. 2019;76(8):1489–1506. doi:10.1007/s00018-019-03005-8
27. Yan M, Wang W, Zhou J, et al. Knockdown of PLAT enhances the anticancer effect of gefitinib in non-small cell lung cancer. J Thorac Dis. 2020;12(3):712–723. doi:10.21037/jtd.2019.12.106
28. Bindea G, Mlecnik B, Tosolini M, et al. Spatiotemporal dynamics of intratumoral immune cells reveal the immune landscape in human cancer. Immunity. 2013;39(4):782–795. doi:10.1016/j.immuni.2013.10.003
29. Gordon SR, Maute RL, Dulken BW, et al. PD-1 expression by tumour-associated macrophages inhibits phagocytosis and tumour immunity. Nature. 2017;545(7655):495–499. doi:10.1038/nature22396
30. Hall M, Liu H, Malafa M, et al. Expansion of tumor-infiltrating lymphocytes (TIL) from human pancreatic tumors. J Immunother Cancer. 2016;4:61. doi:10.1186/s40425-016-0164-7
31. Hwang WT, Adams SF, Tahirovic E, Hagemann IS, Coukos G. Prognostic significance of tumor-infiltrating T cells in ovarian cancer: a meta-analysis. Gynecol Oncol. 2012;124(2):192–198. doi:10.1016/j.ygyno.2011.09.039
32. Paijens ST, Vledder A, de Bruyn M, Nijman HW. Tumor-infiltrating lymphocytes in the immunotherapy era. Cell Mol Immunol. 2021;18(4):842–859. doi:10.1038/s41423-020-00565-9
33. Loi S, Sirtaine N, Piette F, et al. Prognostic and predictive value of tumor-infiltrating lymphocytes in a phase III randomized adjuvant breast cancer trial in node-positive breast cancer comparing the addition of docetaxel to doxorubicin with doxorubicin-based chemotherapy: BIG 02–98. J Clin Oncol. 2013;31(7):860–867. doi:10.1200/jco.2011.41.0902
34. Stanton SE, Disis ML. Clinical significance of tumor-infiltrating lymphocytes in breast cancer. J Immunother Cancer. 2016;4:59. doi:10.1186/s40425-016-0165-6
35. Koch A, Joosten SC, Feng Z, et al. Analysis of DNA methylation in cancer: location revisited. Nat Rev Clin Oncol. 2018;15(7):459–466. doi:10.1038/s41571-018-0004-4
36. Murciano-Goroff YR, Warner AB, Wolchok JD. The future of cancer immunotherapy: microenvironment-targeting combinations. Cell Res. 2020;30(6):507–519. doi:10.1038/s41422-020-0337-2
37. Santoni M, Romagnoli E, Saladino T, et al. Triple-negative breast cancer: key role of tumor-associated macrophages in regulating the activity of anti-PD-1/PD-L1 agents. Biochim Biophys Acta Rev Cancer. 2018;1869(1):78–84. doi:10.1016/j.bbcan.2017.10.007
 © 2021 The Author(s). This work is published and licensed by Dove Medical Press Limited. The full terms of this license are available at https://www.dovepress.com/terms.php and incorporate the Creative Commons Attribution - Non Commercial (unported, v3.0) License.
By accessing the work you hereby accept the Terms. Non-commercial uses of the work are permitted without any further permission from Dove Medical Press Limited, provided the work is properly attributed. For permission for commercial use of this work, please see paragraphs 4.2 and 5 of our Terms.
© 2021 The Author(s). This work is published and licensed by Dove Medical Press Limited. The full terms of this license are available at https://www.dovepress.com/terms.php and incorporate the Creative Commons Attribution - Non Commercial (unported, v3.0) License.
By accessing the work you hereby accept the Terms. Non-commercial uses of the work are permitted without any further permission from Dove Medical Press Limited, provided the work is properly attributed. For permission for commercial use of this work, please see paragraphs 4.2 and 5 of our Terms.