Back to Journals » Cancer Management and Research » Volume 9
Low expression of GATA3 promotes cell proliferation and metastasis in gastric cancer
Authors Wei S, Zhong L, Wang X, Zhang W
Received 1 August 2017
Accepted for publication 5 September 2017
Published 7 December 2017 Volume 2017:9 Pages 769—780
DOI https://doi.org/10.2147/CMAR.S147973
Checked for plagiarism Yes
Review by Single anonymous peer review
Peer reviewer comments 2
Editor who approved publication: Dr Kenan Onel
Shuangqin Wei,1 Liang Zhong,2 Xiaoping Wang,1 Wenju Zhang3
1Department of Gastroenterology, Shanghai University of Medicine and Health Sciences Affiliated Zhoupu Hospital, 2Department of Gastroenterology, Huashan Hospital, Fudan University, 3School of Life Science, Shanghai University, Shanghai, People’s Republic of China
Abstract: GATA3, a member of the GATA zinc finger transcription factor family, has been widely investigated for its role in cancer. Although a recent report has found that GATA3 is downregulated in gastric cancer (GC), the detailed mechanism of GATA3 in GC is still unknown. Here, we investigated whether GATA3 was downregulated in GC patients’ tissue samples and cell lines using quantitative real time polymerase chain reaction and Western blotting. In addition, we conducted several functional experiments to investigate the effect of GATA3 in GC, including cell proliferation, metastasis and epithelial–mesenchymal transition (EMT). The results showed that GATA3 was downregulated in GC tissue samples and cells. Moreover, the expression of GATA3 was associated with tumor size, stage and metastasis. Restoration of GATA3 levels suppressed GC cell proliferation, migration and invasion. Furthermore, chromatin immunoprecipitation and luciferase reporter assay also revealed that GATA3 transcriptionally regulated ZEB1, thereby suppressing EMT. All these findings suggest that GATA3 serves as an oncogene in GC development.
Keywords: invasion, EMT, ZEB1
Introduction
Gastric cancer (GC) has become the fourth most common malignant cancer in the world.1 Almost one million new cases are diagnosed each year.1 Although the treatments, including chemotherapy, radiation therapy and surgery, can increase the survival rate, approximately 60% of patients succumb to GC.2 To improve the prognosis of GC, it is necessary to further decipher the detailed molecular mechanisms of GC progression and development.3–5
GATA zinc finger transcription factor family consists of six members, including GATA-binding protein 1–6. GATA family protein has been found to bind to the specific DNA sequence (A/T)GATA(A/G) and plays an essential role in development, proliferation and differentiation of several types of tissues and cells,6 including the kidney,7 thymocytes,8,9 T-lymphocytes,10 the sympathetic nervous system,11 luminal glandular epithelial cells of the mammary gland,12–14 hair follicles of the skin15 and adipose tissue.16 The significance of this transcription factor has been discussed in recent works, and its role as a tumor suppressor or an oncogene has been debated in many papers.17,18 A low expression of GATA3 has also been found to be associated with a poor prognosis of breast cancer.19,20 Moreover, a recent report showed that GATA3 was downregulated in GC and correlated with poor prognosis in primary gastric adenocarcinoma,21 but the detailed mechanism of GATA3 in GC development is still unknown.
In this work, we found that GATA3 was downregulated in GC tissue samples and cells. Interestingly, the expression of GATA3 was negatively associated with tumor size, stage and metastasis, but there were no correlations between the expression of GATA3 and gender, age and alcohol consumption. The restoration of GATA3 suppressed cell proliferation, migration and invasion. Moreover, GATA3, a transcription factor, transcriptionally regulated ZEB1, thereby inhibiting epithelial–mesenchymal transition (EMT). Therefore, in this study, we proved that GATA3 serves as an oncogene in GC development and acts as a novel therapeutic target.
Materials and methods
Cell culture and collection of tissue samples
Human gastric epithelial cells GES-1 (Bogu Biotech, Shanghai, People’s Republic of China) and human GC cell lines CTC-141 and MKN45 (ATCC, Manassas, VA, USA) were grown in Dulbecco’s Modified Eagle’s Medium (DMEM; HyClone, Logan, UT, USA) supplemented with 100 U/ml penicillin, 100 μg/ml streptomycin and 10% fetal bovine serum (FBS) at 37°C with 5% CO2.
The 83 pairs of GC tissue samples and adjacent normal tissue samples were collected from Zhoupu Hospital during 2013–2016. Written informed consent was obtained from each patient, and the experiments involving human tissue samples were approved by the Clinical Research Ethics Committee of Zhoupu Hospital.
Quantitative real time polymerase chain reaction (qRT-PCR)
RNeasy Mini kit (Thermo Fisher Scientific, Waltham, MA, USA) was used to extract total RNA from tissue samples and cells according to our protocol. Subsequently, cDNA was synthesized using EasyScript Reverse Transcriptase kit (TransGen Biotech, Beijing, People’s Republic of China). qRT-PCR was set up using the TransStart Tip Green qPCR SuperMix (TransGen Biotech) and the targeting gene primers (GATA-3: forward: 5′-GCTTCACAATATTAACAGACCC-3′, reverse: 5′-TTAAACGAGCTGTTCTTGGG-3′; E-cadherin: forward: 5′-ATTTCCCAACTCCTCTCCTG-3′, reverse: 5′-GTTGGATTTGATCTGAACCAGG-3′; α-catenin: forward: 5′-AGGTTATTAATGCTGCACTG-3′, reverse: 5-AATGTGATTCTCTGAGACAGC-3′; N-cadherin: forward: 5′-TCATTAATGAGGGCCTTAAAGC-3′, reverse: 5′-GTTCAGGTAATCATAGTCCTGCT-3′; vimentin: forward: 5′-AAATCCAAGTTTGCTGACCTC-3′, reverse: 5′-GGTCTTGGTATTCACGAAGG-3′; ZEB1: forward: 5′-GGTCTTGGTATTCACGAAGG-3′, reverse 5′-CTTGTCTTTCATCCTGATTTCC-3′). The Ct values were normalized to GAPDH (forward: 5′-TCAAGATCATCAGCAATGCC-3′, reverse: 5′-ATGGACTGTGGTCATGAGTC-3′). Each experiment was performed at least three times.
Western blotting
Tissue samples and cells were collected and washed three times with cold PBS and then homogenized in RIPA lysis buffer (1% NP-40, 0.5% deoxycholic acid, 50 mM Tris-HCl [pH 8.0], 0.1% sodium dodecyl sulfate (SDS), 150 mM NaCl) containing a cocktail of protease inhibitors (Sigma-Aldrich, St Louis, MO, USA). Following sonication and centrifugation (13,000× g, 15 min, 4°C), the supernatant was collected. The BCA protein assay kit (Pierce, Waltham, MA, USA) was used to determine the protein concentration according to our protocol. Samples were denatured in loading buffer, separated by 10% sodium dodecyl sulfate-polyacrylamide gel electrophoresis (SDS-PAGE) and transferred to polyvinylidene fluoride (PVDF) membranes. Anti-GATA-3 (Abcam, Cambridge, UK; diluted 1:200), anti-ZEB1 (Abcam; diluted 1:200), EMT kit (CST, Danvers, MA, USA; diluted 1:1000) and β-actin (Sigma-Aldrich; diluted 1:5000) were incubated at 4°C overnight followed by incubation with HRP-conjugated secondary antibody (Abcam; diluted 1:5000). Specific blots were visualized using the ECL kit (Thermo Fisher Scientific).
Transfection
Cells were transfected with GATA3 siRNA using Lipofectamine RNAi reagent (Thermo Fisher Scientific). The sequences of GATA3 siRNA were: siRNA-1: 5′-GGCUCUACUACA AGCUUCACAAUAU-3′ and siRNA-2: 5′-CCACA CUCUGGAGGAGGAAUGCCAA-3′.22
Cell Counting Kit-8 (CCK-8) analysis
CCK-8 analysis was used to detect the effect of GATA3 on cell proliferation. GATA3 was overexpressed or knocked down in CTC-141 and MKN45 cells. After transcription for 48 h, a total of 3,000 CTC-141 and MKN45 cells were placed in 96-well plates with 200 μl DMEM. Every 24 h, 20 μl CCK-8 regent (Beyotime, Haimen, People’s Republic of China) was added, and cells were incubated at 37°C for 90 min. The absorbance at OD 450 nm revealed the number of surviving cells. Each experiment was performed at least three times.
Colony formation assay
After transfection for 48 h, approximately 5,000 cells were placed in six-well plates and incubated with serum-free DMEM for 12 days, after which cells were washed with PBS three times and fixed with methanol. After staining with 0.1% crystal violet, the number of colonies was counted under a light microscope. Each experiment was performed at least three times.
Cell cycle analysis
CTC-141 and MKN45 cells were transfected with vector, GATA3, scramble siRNA (SCR) or siGATA3,, respectively, for 48 h. Subsequently, cells were collected and fixed with ice-cold 70% ethanol at −20°C overnight. After washing the cells with ice-cold PBS three times, 1 mg/ml RNase A (Sigma-Aldrich) was added, and cells were incubated at 37°C for 60 min. Finally, cells were stained with propidium iodide (Sigma-Aldrich) for 15 min in the dark, and DNA contents were determined using FACScan flow cytometry. Each experiment was performed at least three times.
Transwell migration and invasion assay
Transwell migration and invasion analysis was used to detect the effect of GATA3 on the migratory and invasive behavior of GC cells. Briefly, for Transwell migration and invasion assay, some Transwell chambers (8 μm pore size; EMD Millipore, Billerica, MA, USA) were coated with 100 μl Matrigel (BD, Franklin Lakes, NJ, USA), and some were left uncoated. A total of 4×104 CTC-141 cells were placed on top of Transwell chambers with serum-free DMEM; the lower chambers were filled with 500 μl DMEM containing 10% FBS. Subsequently, cells were incubated at 37°C with 5% CO2 for 18 h, fixed with methanol and stained with 0.1% violet crystal. Cells that remained on the top of the filter were removed using a cotton swab. Finally, the number of migrated and invaded cells was counted under a light microscope. Each experiment was performed at least three times.
Chromatin immunoprecipitation (ChIP) and quantitative chromatin immunoprecipitation (qChIP) assay
Chromatin Immunoprecipitation Assay kit (Beyotime) was used to perform ChIP analysis according to manufacturer’s protocol. Briefly, CTC-141 cells were grown to 80–95% confluence, washed with cold PBS three times and chemically cross-linked with 1% formaldehyde for 40 min at 37°C. Subsequently, cells were lysed with a lysis buffer at 4°C for 60 min and sonicated (four cycles, each cycle 10 times) at 4°C. Following sonication, 2.5 μg GATA3 antibody and anti-rabbit IgG were added to the lysis solution and incubated at 4°C overnight. Protein A beads were used to isolate GATA3- or IgG-interacted DNA fragments. PCR Purification kit (Qiagen, Hilden, Germany) was used to purify the binding chromatin. Each experiment was performed at least three times.
Luciferase reporter assay
To determine whether GATA3 could bind to the promoter region of ZEB1, we cloned a fragment of human ZEB1 promoter harboring the putative GATA3-binding site into the pGL3 Basic vector (Promega, Madison, WI, USA). CTC-141 cells were co-transfected with GATA3 plasmids, and the ZEB1-pGL3 Basic vector was constructed. The dual luciferase assay system (Promega) was used to detect the firefly and Renilla luciferase activities. Each experiment was performed at least three times.
Statistical analysis
All data were analyzed with Statistical Package for Social Sciences software 18.0 (SPSS Inc., Chicago, IL, USA) and expressed as mean±SD. Correlations between groups were determined using two-sided t-tests. One-way analysis of variance together with Dunnett’s multiple comparison tests was used for multiple group comparisons. P<0.05 was considered statistically significant.
Results
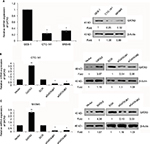
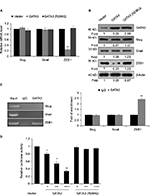
GATA3 is downregulated in GC tissues
A previous study has reported that GATA3 was downregulated in GC.21 To verify this claim, we collected 83 pairs of tumor tissue samples and adjacent normal tissue samples from GC patients. The expression of GATA3 was detected by qRT-PCR and Western blotting. As shown in Figure 1A and B, both mRNA level and protein level of GATA3 were significantly downregulated in tumor tissue samples. To further decipher the role of GATA3 in GC, we analyzed the correlation between the expression of GATA3 and clinical data of GC patients. Interestingly, the expression of GATA3 was found to be associated with tumor size, stage and metastasis, but there were no correlations between the expression of GATA3 and gender, age, pathological grade of cancer and alcohol consumption (Table 1) of patients. We also found that a low expression of GATA2 also predicted a poor prognosis of GC (Figure 1C). Our data confirm that an abnormal expression of GATA3 may play a key role in GC.
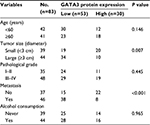  | Table 1 Clinicopathologic variables in 83 gastric cancer patients |
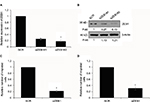
GATA3 is downregulated in GC cells
We also found that GATA3 was dramatically downregulated in several gastric cell lines, such as CTC-141 and MKN45, in comparison to normal human gastric mucosal cell line GES-1 (Figure 2A). We first accomplished an overexpression or knocking down of GATA3 in CTC-141 and MKN45. qRT-PCR and Western blotting were performed to detect the expression of GATA3. As shown in Figure 2B and C, GATA3 was markedly upregulated or knocked down when cells were transfected with FLAG-GATA3 plasmid or GATA3 siRNA, respectively. GATA3 siRNA#1 (siGATA3#1) was found to be more efficient, so it was used for the subsequent experiments.
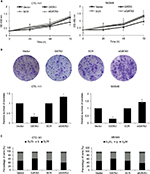
Restoration of GATA3 levels inhibits the proliferation of GC cells
As shown in Table 1, a low expression of GATA3 was associated with tumor size, leading us to assume that a downregulation of GATA3 might facilitate the proliferation of GC cells. To determine the role of GATA3 in cell proliferation, CCK-8 assay and colony formation assay were performed. The results suggested that ectopic expression of GATA3 significantly suppressed the viability of CTC-141 cells compared to control groups; however, an inhibition of GATA3 slightly promoted the viability of CTC-141 cells (Figure 3A). Since the expression level of GATA3 in GC cells is low, the effect of knocking down GATA3 is not obvious. Similarly, an overexpression of GATA3 obviously suppressed the viability of MKN45 cells compared to the control group (Figure 3A). From the colony formation assay, it was confirmed that the restoration of GATA3 levels led to a fall in the number of colonies compared to the control group, but GATA3 inhibition also slightly increased the number of colonies (Figure 3B). We further investigated whether GATA3 suppressed cell proliferation through cell cycle regulation. As shown in Figure 3C, ectopic expression of GATA3 largely increased the percentage of cells in G0/G1 phase and decreased the percentage of cells in S+G2/M phase, compared to the vector group. However, the inhibition of GATA3 decreased the percentage of cells in G0/G1 phase and increased the percentage of cells in S+G2/M phase (Figure 3C). Put together, our work shows evidence that the restoration of GATA3 levels inhibited the proliferation of GC cells.
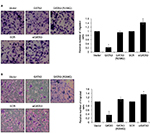
Restoration of GATA3 levels inhibits the migratory and invasive behavior of GC cells
Since a low expression of GATA3 seems to have an association with metastasis, we next detected the effect of GATA3 on cell migration and invasion. Transwell migration and invasion analysis was performed to determine the effect of GATA3 on the migratory and invasive behavior of CTC-141 cells. The results of Transwell migration analysis showed that the migratory behavior of CTC-141 cells that overexpressed GATA3 was obviously reduced compared to the control group, mutant GATA3 (R298Q) had little effect (Figure 4A). However, the migratory behavior of CTC-141 cells that knocked down GATA3 was slightly increased (Figure 4A). Furthermore, Transwell invasion analysis revealed that ectopic expression of GATA3 significantly suppressed the invasive behavior of CTC-141 cells compared to the control groups, mutant GATA3 (R298Q) had little effect; however, inhibition of GATA3 slightly promoted the invasive behavior of CTC-141 cells (Figure 4B). These data suggest that GATA3 suppresses the migration and invasive behavior of GC cells.
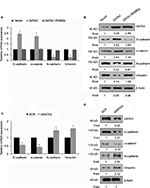
GATA3 inhibits EMT in GC cells
EMT is a complex process which plays a key role in cancer metastasis. Several reports have shown that the GATA family has a significant role to play in EMT.23,24 To further explore the mechanism of GATA3 in GC metastasis, we determined whether GATA3 regulated EMT in GC cells. As shown in Figure 5A and B, the expression of epithelial markers, including E-cadherin and α-catenin, was obviously upregulated in CTC-141 cells that overexpressed GATA3, but the expression of mesenchymal markers, such as N-cadherin and vimentin, was significantly downregulated. Moreover, mutant GATA3 (R298Q) had little effect on EMT (Figure 5A and B). However, GATA3 inhibition suppressed the expression of epithelial markers, including E-cadherin and α-catenin, and promoted the expression of mesenchymal markers, including N-cadherin and vimentin (Figure 5C and D).
GATA3 transcriptionally regulates ZEB1 in GC cells
EMT was found to be regulated by several transcription factors, including slug, snail and ZEB1.25–27 Thus, we next explored whether GATA3 regulated the expression of these transcription factors. As shown in Figure 6A and B, we found that only ZEB1 was regulated by GATA3. We next evaluated whether GATA3 transcriptionally regulated ZEB1. ChIP and qChIP assay revealed that GATA3 bounded with the promoter region of ZEB1 (Figure 6C). Moreover, luciferase reporter assay suggested that wild-type GATA3 transcriptionally suppressed ZEB1, but mutant GATA3 (R298Q) had no effect on ZEB1 (Figure 6D).
Overexpression of ZEB1 suppresses the migratory and invasive behavior of GC cells
We aimed to detect whether GATA3 regulated the migratory behavior of GC cells through the regulation of ZEB1 after transfecting CTC-141 cells with ZEB1 siRNA. As shown in Figure 7A and B, the ZEB1 level was obviously lower in CTC-141 cells. ZEB1 siRNA#2 (siZEB1#2) was found to more efficient, so it was used for the subsequent experiments. Subsequently, Transwell migration and invasion assay was carried out to detect the migratory and invasive behavior of each cell group. Consistent with the effect of GATA3 overexpression, knockdown of ZEB1 also dramatically suppressed the migratory and invasive behavior of CTC-141 cells (Figure 7C and D). These data indicate that GATA3 inhibits the migratory and invasive behavior of GC cells through transcriptionally inhibiting ZEB1.
Discussion
GATA3, a member of the GATA family, has been found to play a key role in facilitating proliferation and development of several tissues and organs.6,17,28,29 In interfollicular epidermis, GATA3 is required during mouse embryonic development.28 A downregulation of GATA3 in breast carcinoma is correlated with poor clinical outcome.12 Therefore, GATA3 may serve as a novel prognostic biomarker.30
In this study, we detected the expression of GATA3 in GC tissues and cells for the first time. Our results suggest that GATA3 was downregulated in GC tissues and cells. Moreover, the expression of GATA3 was negatively correlated with tumor size and metastasis. Furthermore, a low expression of GATA3 predicted poor prognosis. Similar results have been reported in breast cancer studies.19,20 Our work strongly demonstrated that GATA3 plays a significant role in GC development.
Because cell proliferation, migration and invasion are crucial biological features in cancer development, we performed several functional experiments to detect the effect of GATA3 on these biological processes. CCK-8 and colony formation assay indicated that ectopic expression of GATA3 suppressed cell proliferation in GC. In addition, Transwell migration and invasion assay revealed that ectopic expression of GATA3 suppressed cell migration and invasion in GC. Increasing evidence has suggested the important role of EMT in cancer progression, such as metastasis.31,32 So, we explored the effect of GATA3 on EMT in GC cells by measuring the protein level and mRNA level of EMT-associated markers. As we expected, an overexpression of GATA3 obviously suppressed EMT in GC cells through increasing the expression of epithelial markers, including E-cadherin and α-catenin, and decreasing the expression of mesenchymal markers, including N-cadherin and vimentin. Multiple reports have shown that EMT is regulated by several transcription factors, such as snail, slug and ZEB1.23,24 Luciferase reporter assay and ChIP assay revealed that GATA3 transcriptionally regulated ZEB1, thereby inhibiting EMT in GC. It is better to verify our hypothesis in tumor tissue-derived cells, but all tissue samples in our study have been used for performing qRT-PCR and Western blotting analysis, so the effect of GATA3 on tumor tissue-derived cells will be further decipher in our future research.
In conclusion, our work showed that GATA3 was downregulated in GC tissues and cells. The restoration of GATA3 markedly inhibited cell proliferation, migration, invasion and EMT in GC cells. In addition, we found that GATA3 markedly reduced the expression of ZEB1. These findings confirm that GATA3 serves as an oncogene in GC development.
Acknowledgment
This study was supported by Pudong New District Science and Technology Development fund of Shanghai, item number: PKJ2015-Y31.
Disclosure
The authors report no conflicts of interest in this work.
References
Siegel R, Naishadham D, Jemal A. Cancer statistics, 2013. CA Cancer J Clin. 2013;63(1):11–30. | ||
Macdonald JS, Smalley SR, Benedetti J, et al. Chemoradiotherapy after surgery compared with surgery alone for adenocarcinoma of the stomach or gastroesophageal junction. N Engl J Med. 2001;345(10):725–730. | ||
Chen CN, Lin JJ, Chen JJ, et al. Gene expression profile predicts patient survival of gastric cancer after surgical resection. J Clin Oncol. 2005;23(29):7286–7295. | ||
Hippo Y, Taniguchi H, Tsutsumi S, et al. Global gene expression analysis of gastric cancer by oligonucleotide microarrays. Cancer Res. 2002;62(1):233–240. | ||
Oue N, Hamai Y, Mitani Y, et al. Gene expression profile of gastric carcinoma: identification of genes and tags potentially involved in invasion, metastasis, and carcinogenesis by serial analysis of gene expression. Cancer Res. 2004;64(7):2397–2405. | ||
Burch JB. Regulation of GATA gene expression during vertebrate development. Semin Cell Dev Biol. 2005;16(1):71–81. | ||
Grote D, Souabni A, Busslinger M, Bouchard M. Pax 2/8-regulated Gata 3 expression is necessary for morphogenesis and guidance of the nephric duct in the developing kidney. Development. 2006;133(1):53–61. | ||
Pai SY, Truitt ML, Ting CN, Leiden JM, Glimcher LH, Ho IC. Critical roles for transcription factor GATA-3 in thymocyte development. Immunity. 2003;19(6):863–875. | ||
Hendriks RW, Nawijn MC, Engel JD, van Doorninck H, Grosveld F, Karis A. Expression of the transcription factor GATA-3 is required for the development of the earliest T cell progenitors and correlates with stages of cellular proliferation in the thymus. Eur J Immunol. 1999;29(6):1912–1918. | ||
Ting CN, Olson MC, Barton KP, Leiden JM. Transcription factor GATA-3 is required for development of the T-cell lineage. Nature. 1996;384(6608):474–478. | ||
Tsarovina K, Pattyn A, Stubbusch J, et al. Essential role of Gata transcription factors in sympathetic neuron development. Development. 2004;131(19):4775–4786. | ||
Yoon NK, Maresh EL, Shen D, et al. Higher levels of GATA3 predict better survival in women with breast cancer. Hum Pathol. 2010;41(12):1794–1801. | ||
Kouros-Mehr H, Slorach EM, Sternlicht MD, Werb Z. GATA-3 maintains the differentiation of the luminal cell fate in the mammary gland. Cell. 2006;127(5):1041–1055. | ||
Cimino-Mathews A, Subhawong AP, Illei PB, et al. GATA3 expression in breast carcinoma: utility in triple-negative, sarcomatoid, and metastatic carcinomas. Hum Pathol. 2013;44(7):1341–1349. | ||
Kaufman CK, Zhou P, Pasolli HA, et al. GATA-3: an unexpected regulator of cell lineage determination in skin. Genes Dev. 2003;17(17):2108–2122. | ||
Tong Q, Dalgin G, Xu H, Ting CN, Leiden JM, Hotamisligil GS. Function of GATA transcription factors in preadipocyte-adipocyte transition. Science. 2000;290(5489):134–138. | ||
Zheng R, Blobel GA. GATA transcription factors and cancer. Genes Cancer. 2010;1(12):1178–1188. | ||
Takaku M, Grimm SA, Wade PA. GATA3 in breast cancer: tumor suppressor or oncogene? Gene Expr. 2015;16(4):163–168. | ||
Ciocca V, Daskalakis C, Ciocca RM, Ruiz-Orrico A, Palazzo JP. The significance of GATA3 expression in breast cancer: a 10-year follow-up study. Hum Pathol. 2009;40(4):489–495. | ||
Jacquemier J, Charafe-Jauffret E, Monville F, et al. Association of GATA3, P53, Ki67 status and vascular peritumoral invasion are strongly prognostic in luminal breast cancer. Breast Cancer Res. 2009;11(2): R23. | ||
Keshari RP, Wang W, Zhang Y, et al. Decreased expression of the GATA3 gene is associated with poor prognosis in primary gastric adenocarcinoma. PLoS One. 2014;9(2):e87195. | ||
Liu X, Cao K, Xu C, et al. GATA-3 augmentation down-regulates Connexin43 in Helicobacter pylori associated gastric carcinogenesis. Cancer Biol Ther. 2015;16(6):987–996. | ||
Martinelli P, Carrillo-de Santa Pau E, Cox T, et al. GATA6 regulates EMT and tumour dissemination, and is a marker of response to adjuvant chemotherapy in pancreatic cancer. Gut. 2017;66(9):1665–1676. | ||
Campbell K, Whissell G, Franch-Marro X, Batlle E, Casanova J. Specific GATA factors act as conserved inducers of an endodermal-EMT. Dev Cell. 2011;21(6):1051–1061. | ||
Deng Y, Luan F, Zeng L, Zhang Y, Ma K. MiR-429 suppresses the progression and metastasis of osteosarcoma by targeting ZEB1. EXCLI J. 2017;16:618–627. | ||
Fan F, Samuel S, Evans KW, et al. Overexpression of snail induces epithelial-mesenchymal transition and a cancer stem cell-like phenotype in human colorectal cancer cells. Cancer Med. 2012;1(1):5–16. | ||
Toiyama Y, Yasuda H, Saigusa S, et al. Increased expression of Slug and Vimentin as novel predictive biomarkers for lymph node metastasis and poor prognosis in colorectal cancer. Carcinogenesis. 2013;34(11):2548–2557. | ||
de Guzman Strong C, Wertz PW, Wang C, et al. Lipid defect underlies selective skin barrier impairment of an epidermal-specific deletion of Gata-3. J Cell Biol. 2006;175(4):661–670. | ||
Pandolfi PP, Roth ME, Karis A, et al. Targeted disruption of the GATA3 gene causes severe abnormalities in the nervous system and in fetal liver haematopoiesis. Nat Genet. 1995;11(1):40–44. | ||
Chou J, Provot S, Werb Z. GATA3 in development and cancer differentiation: cells GATA have it! J Cell Physiol. 2010;222(1):42–49. | ||
Takai M, Terai Y, Kawaguchi H, et al. The EMT (epithelial-mesenchymal-transition)-related protein expression indicates the metastatic status and prognosis in patients with ovarian cancer. J Ovarian Res. 2014;7:76. | ||
Hong H, Zhou T, Fang S, et al. Pigment epithelium-derived factor (PEDF) inhibits breast cancer metastasis by down-regulating fibronectin. Breast Cancer Res Treat. 2014;148(1):61–72. |
 © 2017 The Author(s). This work is published and licensed by Dove Medical Press Limited. The full terms of this license are available at https://www.dovepress.com/terms.php and incorporate the Creative Commons Attribution - Non Commercial (unported, v3.0) License.
By accessing the work you hereby accept the Terms. Non-commercial uses of the work are permitted without any further permission from Dove Medical Press Limited, provided the work is properly attributed. For permission for commercial use of this work, please see paragraphs 4.2 and 5 of our Terms.
© 2017 The Author(s). This work is published and licensed by Dove Medical Press Limited. The full terms of this license are available at https://www.dovepress.com/terms.php and incorporate the Creative Commons Attribution - Non Commercial (unported, v3.0) License.
By accessing the work you hereby accept the Terms. Non-commercial uses of the work are permitted without any further permission from Dove Medical Press Limited, provided the work is properly attributed. For permission for commercial use of this work, please see paragraphs 4.2 and 5 of our Terms.