Back to Journals » Breast Cancer: Targets and Therapy » Volume 9
Loss of constitutive ABCB1 expression in breast cancer associated with worse prognosis
Authors Delou JM, Vignal GM, Índio-do-Brasil V, Accioly MT, da Silva TS, Piranda DN, Sobral-Leite M, Carvalho MA , Capella MA, Vianna-Jorge R
Received 29 December 2016
Accepted for publication 22 March 2017
Published 10 June 2017 Volume 2017:9 Pages 415—428
DOI https://doi.org/10.2147/BCTT.S131284
Checked for plagiarism Yes
Review by Single anonymous peer review
Peer reviewer comments 3
Editor who approved publication: Professor Pranela Rameshwar
João Marcos de Azevedo Delou,1–3 Giselle Maria Vignal,4 Vanessa Índio-do-Brasil,2,5 Maria Theresa de Souza Accioly,4 Taiana Sousa Lopes da Silva,2 Diogo Nascimento Piranda,2,3 Marcelo Sobral-Leite,2,6 Marcelo Alex de Carvalho,2,7 Márcia Alves Marques Capella,1,8 Rosane Vianna-Jorge2,3,5
1Programa de Bioquímica e Biologia Celular, Instituto de Bioquímica Médica, Universidade Federal do Rio de Janeiro, 2Programa de Farmacologia, Coordenação de Pesquisa, Instituto Nacional de Câncer, 3Instituto de Ciências Biomédicas, Universidade Federal do Rio de Janeiro, 4Divisão de Patologia, Instituto Nacional de Câncer, 5Escola Nacional de Saúde Pública – FIOCRUZ, Rio de Janeiro, Brazil; 6Department of Molecular Pathology, Netherlands Cancer Institute, Amsterdam, The Netherlands; 7Instituto Federal do Rio de Janeiro, 8Instituto de Biofísica Carlos Chagas Filho, Universidade Federal do Rio de Janeiro, Rio de Janeiro, Brazil
Abstract: ABCB1 gene encodes an adenosine 5’-triphosphate–binding cassette transporter, which not only confers multidrug resistance phenotype in malignant cells, but is also present in several nonmalignant tissues. For the last thirty years, ABCB1 expression in breast cancer has been described by many authors, but the extent of expression differs among the studies, and there is no consensus regarding its potential role in carcinogenesis or in the tumor response to antineoplastic drugs. This study aimed to characterize the expression of ABCB1 in breast tumors as a function of genetic, clinical, and histopathological variables. The ABCB1 expression was also evaluated in nonmalignant mammary tissues adjacent to tumors and in benign lesions. The detection of ABCB1 protein was performed by immunohistochemistry in tissue specimens of excised breasts obtained from a prospective cohort of Brazilian women with breast cancer. The association of ABCB1 protein levels with ABCB1 mRNA, gene polymorphisms, and clinical and histopathological variables was also evaluated. The Kaplan–Meier curves and multivariate Cox regression analyses were conducted to identify independent predictors of disease-free survival of patients with breast cancer. ABCB1 was detected in 86.3% (656) of breast tumors, 98.8% (606) of nonmalignant mammary tissue adjacent to tumors, and 100% (28) of benign lesions. Reduced ABCB1 protein levels in breast tumors was associated with triple-negative subtype (adjusted odds ratio [ORadj] =0.24; 95% confidence interval [CI] =0.13–0.45), lymph node status < pN2 (ORadj =0.27; 95% CI =0.10–0.71), tumor size >2 cm (ORadj =0.55; 95% CI =0.32–0.93), and hypertensive status (ORadj =0.42; 95% CI =0.24–0.73), and it was significantly associated with shorter disease-free survival, either for all breast cancer patients (p log-rank =0.012; hazard ratio [HR] =3.46; 95% CI =1.21–9.91) or for those with triple-negative tumors (p log-rank =0.007; HR =11.41; 95% CI =1.29–100.67). The loss of constitutive ABCB1 expression in breast cancer, especially in triple-negative tumors, seems to indicate a subgroup of worse prognosis.
Keywords: multidrug resistance, single-nucleotide polymorphisms, immunohistochemistry, disease-free survival, triple-negative breast cancer, hypertension
Introduction
Efflux of cytotoxic drugs by the adenosine 5’-triphosphate–binding cassette transporter subfamily B member 1 (ABCB1 aka multidrug resistance protein 1 [MDR1]/P-glycoprotein) is considered a potential mechanism of acquired chemoresistance to antineoplastics.1 However, there is a controversy regarding the role of ABCB1 expression in breast cancer.2,3 Literature discrepancies may involve the lack of standardized methods for the detection and quantification of ABCB1 in solid tumors.4 For instance, there is a long-lasting notion of uncertainty regarding ABCB1 detection due to the lack of sensitivity and/or specificity of several commercial ABCB1 antibodies.2,5,6 In addition, there seems to be great interpatient variability.3 One possible reason for such variability is that the ABCB1 gene is polymorphic, and two of its single-nucleotide polymorphisms (SNPs), rs1128503 and rs1045642, may modify the final protein conformation, compromising its membrane stability and substrate recognition.7,8 Finally, it has been reported that the ABCB1 expression may also be modulated by plasma aldosterone9 or cortisol,10 as well as by dietary salt and dehydration.9
In this study, a systematic evaluation of ABCB1 expression in the breast, encompassing benign lesions, breast tumors examined prior to any chemotherapeutic treatment and non-malignant mammary tissues adjacent to breast tumors, was done. The study was conducted in a prospective manner by using the tissue samples from a cohort of 712 Brazilian women who underwent curative mammary surgery. ABCB1 expression was estimated by immunohistochemistry with three previously validated antibodies, and its association with clinical and histopathological variables, including the genetic profile regarding ABCB1 SNP, was evaluated. Finally, the impact of ABCB1 expression on short-time (2-year) disease-free survival of patients with breast cancer was also investigated.
Methods
Subjects and study design
The study population was a prospective cohort of Brazilian women who were admitted from January 2009 to December 2012 at Instituto Nacional de Câncer (INCA) for mammary surgery. The exclusion criteria were the following: any previous oncological treatment, prior contralateral or bilateral synchronous breast cancer, and systemic metastasis at diagnosis. Figure 1 illustrates the flowchart of the study design, depicting the reasons for exclusion and the sample availability for each analysis.
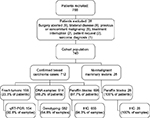  | Figure 1 Flowchart of the study cohort. Abbreviations: qRT-PCR, quantitative real-time polymerase chain reaction; IHC, immunohistochemistry. |
The study protocol (approved by the Ethics Committee of INCA #129/08) did not interfere with the routine clinical follow-up or therapeutic choice. All the patients provided written informed consent to be enrolled in the present study. The REMARK guidelines for the characterization of biomarkers11 and the international precepts of ethics in research and of good clinical practice were followed.
Clinical and histopathological characterization
A description of this study cohort has been published previously.12 All the patients were interviewed to provide information on their clinical history and lifestyle habits. The variables considered for clinical history were age at diagnosis, menopausal status, and comorbidities, which were defined as any preexisting chronic condition under medical treatment, with the exception of obesity, which was defined based on the body mass index. Hypertension was defined according to disease severity, as inferred by the prescribed antihypertensive therapy.13 The lifestyle variables were alcohol drinking, defined according to the frequency of consumption, and smoking, classified as current, previous, or no habit, considering a minimum consumption of five packs (100 cigarettes).
Histopathological characterization of breast tumors was based on the 3rd edition of the World Health Organization Classification of Tumors14 and on the Elston–Ellis histological grading system.15 Data on hormone receptors and human epidermal growth factor receptor 2 (HER-2) status of breast tumors, according to immunohistochemical and fluorescence in situ hybridization analyses, were used for the surrogate classification of tumor subtypes.16
Immunohistochemistry, antibody validation, and scoring
All paraffin-embedded blocks were stained with hematoxylin and eosin to select the most representative specimen for each patient, which was processed as described previously.17 ABCB1 detection was tested with three antihuman ABCB1 monoclonal antibodies, namely sc-13131, clone G-1 (Santa Cruz Biotechnology Inc, Dallas, TX, USA.); ab3083, clone 265/F4 (Abcam, Cambridge, UK); and NCL-PGLYm, clone 5B12 (Leica Biosystems Newcastle Ltd., Newcastle, UK), and then revealed with Novolink Polymer Detection System standard protocol (Leica Biosystems Newcastle Ltd.). The most sensitive antibody among these three was sc-13131, clone G-1 (dilution 1:10,000).
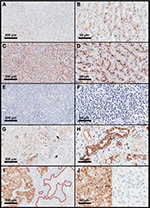
Although the G-1 specificity toward ABCB1 has already been shown,18–21 validation experiments were conducted in this study, as recommended by the consensus for ABCB1 detection,2,5 using specimens from different nonmalignant human tissues. Liver, adrenal, and kidney proximal and distal tubules were used as positive controls,22–24 whereas tonsils, epididymis, and kidney glomeruli were used as negative controls.23–25 Because ABCB1 was expected to be detected in the biliary canaliculi,23 liver samples were also stained with anti-CD10 antibody (clone 56C6; Leica Biosystems Newcastle Ltd.; diluted 1:200). Liver samples were positive for ABCB1 (Figure 2A and B), with a distribution pattern similar to that observed with the anti-CD10 antibody (Figure 2C and D). The cross-reaction against ABCB4/MDR3 was investigated using human tonsils and kidney. Tonsils and kidney glomeruli showed no immunostaining (Figure 2E–G [arrows]), indicating the absence of G-1 antibody cross-reaction toward ABCB4 (formerly known as MDR3).26,27 Proximal and distal kidney tubules, which express ABCB1,24 showed positive staining (Figure 2G and H), whereas kidney glomeruli (arrows in Figure 2G), which express only ABCB4,27 were negative (Figure 2G and H), confirming the absence of cross-reactivity against ABCB4/MDR3.
Because of the observed intratumoral variability in ABCB1 staining in breast tumors (Figure 2I and J), the individual quantification included the whole area of a representative tumor slide and was based on the two previously published scoring methods: a continuous scale (CS)28 and a categorical score,29 both of which consider the percentage of immunostained cells and the intensity of the reaction.
The CS was calculated as follows: CS = (%weak × 1) + (%moderate × 2) + (%strong × 3).28 The intensity of ABCB1 staining was rated as follows: negative (complete absence of cellular reaction), weak (diffuse and mild reaction in cytoplasm, with no detectable reaction in cell membranes), moderate (detectable reaction in both cytoplasm and plasma membrane), or strong (strong in both cytoplasm and plasma membrane). Nuclear reaction was not considered.
The categorical score, or immunoreaction score (IRS) was defined by the following equation: IRS = (positivity score) × (intensity score).29 The positivity score was attributed 1 to 4, according to the percentage of positive cancer cells: 1 (1%–9%), 2 (10%–49%), 3 (50%–79%), or 4 (80%–100%). The intensity score ranged 0–3: negative (0), weak (1), moderate (2), or strong (3). Breast tissues were considered positive for ABCB1 when the IRS was ≥4, meaning that at least 10% of the cells presented moderate staining.29
All the slides were blindly evaluated by a trained PhD student (JMAD) and a pathologist (GMV).
Genotyping analyses
Genomic DNA was extracted from peripheral blood samples (3 mL) by using the Blood Genomic Prep Mini Spin Kit (GE Heathcare, Buckinghamshire, UK). Genotyping analyses were conducted by using TaqMan real-time polymerase chain reaction (RT-PCR) assays in an ABI PRISM 7500 Sequence Detector System (Applied Biosystems, Foster City, CA, USA). Each reaction contained 1 µL of genomic DNA (20 ng/µL); 0.5 µL of probe, either C_7586662_10 for rs1128503 or C__7586657_20 for rs1045642 (Applied Biosystems); 5µL of Genotyping Master Mix (Applied Biosystems); and 3.5 µL of water. All the experiments were carried out in 96-well plates, including a nontemplate control and at least two positive controls.
Quantification of ABCB1 mRNA
Fresh specimens of excised breast tumors were dissected by clinical pathologists, frozen in liquid N2, and stored at Banco Nacional de Tumores (BNT/INCA). Frozen sections of breast tumors were used for RNA isolation by using the RNeasy Mini Kit (Qiagen, Valencia, CA, USA). The RNA samples were stored in RNase-free distilled water at −80°C, and the cDNA was synthesized by using 2 μg of RNA, with High-Capacity cDNA Reverse Transcription Kit (Applied Biosystems), according to the manufacturer’s instructions.
The relative quantification (RQ) of ABCB1 transcripts was performed by using by using TaqMan quantitative real-time PCR (qRT-PCR) assays, in an ABI PRISM 7500 Sequence Detector System (Applied Biosystems). Each reaction contained cDNA templates (40 ng), 10 μL of reaction mix containing 5 μL TaqMan Gene Expression Master Mix, and TaqMan probes (Applied Biosystems), which were as follows: ABCB1 Hs01067802_m1* (with FAM), peptidylprolyl isomerase A (PPIA) Hs00216455_ml (with VIC®), used as reference gene. The thermal cycling conditions comprised an initial denaturation step at 95°C for 10 minutes, followed by 40 cycles of 95°C denaturation for 15 seconds, and annealing at 60°C for 1 minute. The experiments were carried out in 96-well plates, including a nontemplate control and a reference sample, consisting of cDNA obtained from a commercial human mammary gland (HMG) total RNA (Clontech Laboratories, Mountain View, CA, USA). The RQ of ABCB1 mRNA was calculated as follows: RQ = 2-∆∆Ct, where ∆∆Ct = ∆CtABCB1 - ∆CtHMG, with ∆CtABCB1 = CtABCB1 - CtPPIA and ∆CtHMG = CtHMG - CtPPIA. All the data were generated in triplicates and expressed as median with their respective 95% confidence intervals (CI).
Characterization of outcomes in patients with breast cancer
Breast cancer progression was characterized by locoregional or contralateral recurrence or distant metastasis. New primary cancer lesions or deaths unrelated to breast cancer progression were censored. The patients were considered disease-free if they had no imaging diagnosis of disease progression or suggestive clinical symptoms up till their last medical consult. Because this is an ongoing cohort, the survival analysis was limited to 2 years of follow-up, which was available for all the patients.
Statistical analyses
A descriptive study was conducted. Clinical and histopathological variables were categorized and expressed as number and relative frequencies. The association between IRS and ABCB1 genotypes and clinical or histopathological variables was evaluated by using the χ2 test. Individual variables were tested for linear-by-linear associations, with calculation of trend significances (ptrend<0.05) and categorized for better or worse prognosis, with the calculation of the odds ratios (ORs) and respective 95% CI. These variables were also compared for the continuous scale by using Mann–Whitney U test or Kruskal–Wallis test. Wald χ2 test was used to identify independent predictors (p<0.05), which were used for the calculation of the corresponding adjusted ORs (ORadj). The final regression model was tested with the Hosmer–Lemeshow test. All the statistical analyses were conducted by using IBM SPSS Version 20 for Windows (IBM Corp., Armonk, NY, USA) or GraphPad Prism 5.0 (GraphPad Software, La Jolla, CA, USA).
Disease-free survival curves were estimated by using the Kaplan–Meier product-limit method, and the impact of the individual variables was estimated by using log-rank analysis (p<0.05). The significant covariates were included in multivariate Cox proportional hazards regression with the calculation of adjusted hazard ratios (HRadj) and respective 95% CIs.
Survival analyses were also performed by using ABCB1 expression data obtained from publically available gene expression array databases. Two web platforms were evaluated: ProgGeneV2.0,30 which comprises several independent breast cancer cohorts (only cohorts with at least 200 breast cancer patients were selected), and the Gene Expression Omnibus deposited data (https://www.ncbi.nlm.nih.gov/geo/), which was assessed via the online software KM plotter (www.kmplot.com),31 by using the filter for breast cancer and information regarding ABCB1. In both the platforms, the median value of ABCB1 mRNA was used as a cutoff value to categorize ABCB1 tumoral expression as “low” or “high”.
Results
Characterization of patients with breast cancer
Table 1 presents the main clinical and histopathological characteristics of the breast cancer patients (n=712) and the allelic and genotypic distribution of rs1128503 (ABCB1 C1236T) and rs1045642 (ABCB1 C3435T).
After surgery, most patients received adjuvant protocols, including chemotherapy (61.4%), hormonal therapy (12.9%), hormonal therapy plus radiotherapy (10.8%), or radiotherapy alone (5.8%). The remaining 9.1% of the patients were clinically followed up with no secondary intervention. The main protocol for adjuvant chemotherapy, comprising 89% of cases, was three cycles of cyclophosphamide, doxorubicin, and 5-fluorouracil, followed by three cycles of docetaxel.32,33
Immunohistochemical characterization of ABCB1 in breast cancer
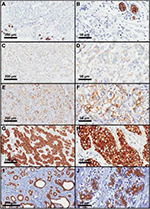
Figure 3 shows ABCB1 immunostaining in the breast, including breast tumors (Figure 3A–H) and normal mammary tissues from women without breast cancer (Figure 3I and J). Figure 3A and B illustrates fully negative reactions, whereas Figure 3C–H shows a gradation of the immunostaining, which was graded as weak (Figure 3C and D), moderate (Figure 3E and F), or strong (Figure 3G and H). Nonmalignant mammary tissue adjacent to the tumor area (n=606) showed positive reaction for ABCB1 in mammary ducts and acini (98.8% of cases), even when tumor staining was negative (Figure 3A and B). The absence of ABCB1 in both the cancer cells and adjacent non-malignant breast tissue occurred in only five patients (0.8% of all tumor samples). All the cases of benign lesions (n=28) were homogeneously positive for ABCB1, with strong immunostaining in all mammary ducts and acini.
Figure 4 shows the distribution of ABCB1 immunostaining scores in the cohort. Figure 4A presents a bar graph of the IRSs, whereas Figure 4B is a histogram of the CS. According to the IRSs, 86.3% of breast tumors were positive for ABCB1 (IRS ≥4), and 71% presented high immunostaining scores (IRS ≥8). The histogram of the CS does not fit into a normal distribution (p<0.001; D’Agostino–Pearson normality test), and 71% of tumors had high CS values (≥150).
Association between ABCB1 mRNA and ABCB1 protein levels in breast tumors
Figure 5 depicts the association between ABCB1 mRNA and ABCB1 protein levels in breast tumors. The stratification of tumors according to their IRSs, as positive or negative for ABCB1, indicates a significant difference in the levels of ABCB1 mRNA (Figure 5A), which were, on average, ~2 times higher among ABCB1 positive tumors. However, the analysis of individual data on ABCB1 mRNA and on ABCB1 protein levels (evaluated as CS) indicates no linear correlation (Figure 5B) and points to a large variability in the distribution of ABCB1 mRNA in breast tumors. In order to explore the possible causes of such variability, the influence of ABCB1 SNPs was investigated. Figure 5C indicates a significant decrease in ABCB1 mRNA levels among carriers of the variant homozygous ABCB1 1236TT (rs1128503), as compared to the other two groups of genotypes. The SNP rs1045642, however, showed no influence on the amounts of ABCB1 mRNA in breast tumors (Figure 5D).
Influence of clinical and histopathological parameters on ABCB1 expression in breast tumors
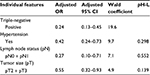
Patients with positive or negative ABCB1 immunostaining in breast tumors were compared with regard to clinical, genetic, and histopathological variables. The conditions significantly associated with negative ABCB1 were hypertensive status, tumor size >2 cm, low lymph node status (pN0 or pN1), early tumor stage (I or II), negative estrogen receptor (ER) status, negative progesterone receptor (PR) status, and triple-negative subtype (Table 2).
According to the final multivariate model (Table 3), hypertensive status, large tumor size, low lymph node status, and triple-negative subtype were independently associated with ABCB1 negativity.
Because all hypertensive patients were under pharmacological treatment, the effect of antihypertensive drugs on ABCB1 immunostaining was investigated (Table 4). No significant associations were found for individual drugs or their pharmacological groups. However, ABCB1 positivity was lower in more severe cases (ptrend<0.001).
Influence of decreased ABCB1 expression on disease-free survival of patients with breast cancer
Figure 6 shows the influence of ABCB1 expression on the 2-year disease-free survival of patients with breast cancer. The results indicated that the loss of ABCB1 expression (IRS ≤4) favors early-onset breast cancer progression when evaluated for all the patients in the cohort (Figure 6A) or for patients with triple-negative tumors (Figure 6B). The impact of the loss of ABCB1 expression on the rates of breast cancer progression was maintained after adjustment for other prognostic factors, that is, HER-2-like or triple-negative subtypes, staging group ≥II, histological grade 3, and the use of chemotherapy (HR =3.5; 95% CI =1.2–9.9) when all the tumors were considered; or staging group ≥II and histological grade 3 for triple-negative tumors (HR =11.4; 95% CI =1.3–100.7).
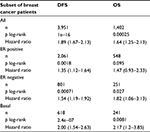
The influence of ABCB1 expression on the 2-year disease-free survival of patients with breast cancer was also evaluated, considering ABCB1 mRNA levels in tumors that could be fresh-frozen at the time of the excision surgery (n=154). A significant difference was detected (p log-rank =0.012), with the patients whose tumors had low ABCB1 mRNA presenting worse cumulative survival than the patients with high ABCB1 mRNA (Figure 6C). However, because of the lack of events in the group with high mRNA, the calculation of the HR was compromised. Therefore, the survival analysis based on ABCB1 mRNA was extended to publically available databases from breast cancer cohorts (Tables 5 and 6). The results obtained with these cohorts confirmed that low ABCB1 mRNA is associated with worse survival outcomes (both disease-free survival and overall survival), either when considered for all the patients with adjustment for ER positivity or when evaluated only among ER negative subtype or among the basal subtype (Table 6).
Finally, the influence of ABCB1 SNPs on the 2-year disease-free survival was also evaluated, but no significant association with early-onset breast cancer progression was detected either for rs1128503 (p log-rank =0.57) or for rs1045642 (p log-rank =0.55).
Discussion
The present work characterized the ABCB1 expression in breast cancer in comparison with nonmalignant breast tissue. All methodological recommendations for the detection of ABCB1 by immunohistochemistry2,4 were followed. First, three different antibody clones (ie, G-1, 5B12, 265/F4) were tested. The most sensitive clone was G-1, whose specificity towards ABCB1 was confirmed (Figure 2). Second, breast tumors were processed immediately after resection, by using buffered formalin. Third, a polymer-based kit was used to improve antibody sensitivity.42 Fourth, two different scoring methods28,29 were used. Finally, ABCB1 mRNA was quantified in freshly frozen tumors, testing a second parameter of ABCB1 expression. In addition, the study was conducted in a prospective manner, by using all the available samples from a relatively large cohort of patients (656 tissue samples from 712 patients). For comparison, a literature review on ABCB1 detection by immunohistochemistry in breast cancer retrieved only nine studies with ≥100 patients.17,29,43–49
The results on ABCB1 expression in breast tumors indicated high interindividual variability, considering both the percentage of immunostained cells and the intensity of the immunostaining reaction. A meta-analysis conducted by Trock et al,2 comprising 31 studies, also indicated a high heterogeneity in ABCB1 positivity, with a weighted average value of 48.5% (95% CI =42–55.0), and a trend for higher values in more recent studies. For example, in two recent retrospective studies, the reported ABCB1 detection was 42% in 104 patients29 or 66% in 177 patients.48
The evaluation of nonmalignant mammary tissues indicated the presence of ABCB1 in all cases of benign breast lesions, as well as in the normal ducts and acini of 98.8% of breast cancer samples. Such results are in agreement with the findings of Pavelic et al,50 who described ABCB1 immunoreactivity in normal ductal epithelia using four independent antibodies. Likewise, Scala et al51 showed that >80% of normal breast ductal epithelium stained positively for ABCB1, with staining being confined to the luminal surface.51 Considering that most breast tumors have a ductal origin, it seems that ABCB1 detection in breast carcinoma is a consequence of constitutive ABCB1 expression in the breast, rather than an acquired or rare phenotype. However, Zhu et al52 found higher ABCB1 expression in breast cancer tissues (57.3%) compared with adjacent noncancerous tissues (5.0%).
As an attempt to investigate the individual aspects that may modulate ABCB1 expression in breast tumors, mRNA levels and ABCB1 genotypes were evaluated. ABCB1 mRNA levels were reduced in tumors from patients carrying the TT genotype of rs1128503, although no significant effect was detected in the final protein levels. There are no previous reports on the effect of rs1128503 in breast tumors, but a recent review indicated no effect on ABCB1 protein levels or activity in acute myeloid leukemia.53 With regard to rs1045642, previous studies suggest that the TT genotype might lead to decreased ABCB1 mRNA levels in mammary carcinoma cell lines,54 or in breast tumor,55 which was not confirmed by the present study . Taken together, these results highlight the risks of using mRNA quantification to infer protein levels or activity. Accordingly, there seems to be a posttranscriptional regulation of ABCB1 expression, possibly mediated by microRNAs.56
The evaluation of ABCB1 immunostaining according to histopathological variables indicated lower expression among tumors with large size, low lymph node status, or triple-negative subtype. Some other authors also found positive associations between ABCB1 in breast tumors and lymph node metastases29,52,57,58 or positive ER status.43,45 However, Kuroda et al49 found higher ABCB1 immunostaining with the reduction or loss of ER, PR, and HER-2. They identified ABCB1 in 29 (59.2%) of 49 basal-like carcinomas, characterized by the presence of cytokeratins 5/6, 14, or 17, as compared to 85 (30.6%) among 278 non-basal-like carcinomas. In the present study, the expression of cytokeratins was not evaluated. Thus, ABCB1 expression according to the luminal or basal origin of breast carcinoma or among the triple-negative basal-subtypes could not be inferred.
With regard to the association of decreased ABCB1 expression with hypertension, an experimental model of spontaneously hypertensive rats suggested lower ABCB1 activity in the kidneys and in peripheral blood mononuclear cells.59 In addition, ABCB1 protein levels were also diminished in the kidneys of rats submitted to high-sodium diet9,60 or to adrenalectomy.9 Because a mineralocorticoid receptor61 and a tissue renin–angiotensin system62 have been identified in normal and malignant breast tissues, it is possible that this pathway may modulate ABCB1 expression in breast cancers and that a disruption of the renin–angiotensin system in the malignant tissue62 may contribute to lower ABCB1 expression in some cases.
Finally, the analysis of breast cancer outcomes suggested that the loss of ABCB1 expression is associated with early-onset disease progression, especially among patients with triple-negative tumors. There is no presumed causal mechanism to justify how the loss of ABCB1 in breast cancer would favor cell survival, proliferation, migration, or invasion. Instead, the recognized actions of ABCB1 as a physiological regulator of cell cycle and apoptosis and of the lipid turnover and maintenance of membrane structure are expected to protect cancer cells from death.63 It is believed that the loss of ABCB1 in breast cancer tissue might be a consequence of the genomic instability in cancer cells, especially among triple-negative tumors, which also lack other regulatory proteins, including the ER and HER-2, rather than a specific adaptation leading to a more aggressive phenotype.
The results obtained with publically available databases corroborated the findings of the present study and suggested that low ABCB1 mRNA may predict shorter disease-free survival and overall survival even when considering only patients with triple-negative clusters, such as the basal-type, which poses the greatest challenge for treatment because of its aggressive clinical course and lack of targeted therapy.64 Taken together, the current results indicated that negative or low ABCB1 expression in breast tumors, rather than being beneficial due to a possible reduction of drug efflux, seems to indicate a more aggressive phenotype, which may occur in luminal tumors, but is more frequent among triple-negative tumors.
Conclusion
In conclusion, the study results point to important paradigm changes in the concept of chemoresistance in breast cancer: 1) ABCB1 seems to be constitutively expressed in the normal mammary tissue, being maintained, rather than acquired, in nonmalignant lesions and in most cases of breast cancer; 2) ABCB1 protein levels might be downregulated in some breast cancers, especially in triple-negative tumors, or as a consequence of systemic arterial hypertension, although the prognostic impact of such association is yet to be determined; and 3) the absence of ABCB1 in triple-negative tumors might contribute to identify a subgroup with worse prognosis.
Acknowledgments
The authors thank Dr Guilherme Suarez-Kurtz for the use of laboratory facilities and the personnel from the Breast Cancer Hospital, from the Division of Pathology, and from the National Bank of Tumors in the Brazilian National Cancer Institute for logistic support in sample and data collection. This study was supported by grants from Conselho Nacional de Desenvolvimento Científico e Tecnológico (CNPq 474522/2010-5), Fundação Carlos Chagas Filho de Amparo à Pesquisa do Estado do Rio de Janeiro (FAPERJ E-26/110356/2010), and Instituto Nacional de Ciência e Tecnologia para o Controle do Câncer (CNPq 573806/2008-0; FAPERJ E26/170.026/2008).
Author contributions
JMAD recruited patients, collected clinical and histopathological data, characterized genotypes and haplotypes, helped evaluating patients’ blocks and slides, conducted immunohistochemical and mRNA quantification assays, performed all statistical analyses, generated tables and figures. GMV evaluated surgery resections, selected blocks, and evaluated patients’ slides. VI-do-B recruited patients, collected clinical information, and helped with the statistical analyses. MTSA coordinated the immunohistochemical analyses. TSLS helped recruiting patients and set the genotyping assays. DNP set and performed mRNA expression assays. MSL performed mRNA expression assays and collected histopathological data. MAMC and MAC coordinated mRNA expression assays. RVJ conceived, designed, and coordinated the study; analyzed the data; all authors contributed toward data analysis, drafting and revising the paper and agree to be accountable for all aspects of the work.
Disclosure
JMAD received a PhD scholarship from Conselho Nacional de Desenvolvimento Científico e Tecnológico (CNPq 140789/2009-0). DNP and MSL received graduate scholarships from Coordenação de Aperfeiçoamento de Pessoal de Nível Superior, and TSLS received an undergraduate scholarship from CNPq. The other authors report no conflicts of interest in this work.
References
Martin HL, Smith L, Tomlinson DC. Multidrug-resistant breast cancer: current perspectives. Breast Cancer (Dove Med Press). 2014;6:1–13. | ||
Trock BJ, Leonessa F, Clarke R. Multidrug resistance in breast cancer: a meta-analysis of MDR1/gp170 expression and its possible functional significance. J Natl Cancer Inst. 1997;89(13):917–931. | ||
Wind NS, Holen I. Multidrug resistance in breast cancer: from in vitro models to clinical studies. Int J Breast Cancer. 2011;2011:967419. | ||
Beck WT, Grogan TM, Willman CL, et al. Methods to detect P-glycoprotein-associated multidrug resistance in patients’ tumors: consensus recommendations. Cancer Res. 1996;56(13):3010–3020. | ||
Clarke R, Leonessa F, Trock B. Multidrug resistance/P-glycoprotein and breast cancer: review and meta-analysis. Semin Oncol. 2005;32(6 Suppl 7):S9–S15. | ||
Cianfriglia M. The biology of MDR1-P-glycoprotein (MDR1-Pgp) in designing functional antibody drug conjugates (ADCs): the experience of gemtuzumab ozogamicin. Ann Ist Super Sanità. 2013;49(2):150–168. | ||
Kimchi-Sarfaty C, Oh JM, Kim I, et al. A “silent” polymorphism in the MDR1 gene changes substrate specificity. Science. 2007;315(5811):525–528. | ||
Fung KL, Pan J, Ohnuma S, et al. MDR1 synonymous polymorphisms alter transporter specificity and protein stability in a stable epithelial monolayer. Cancer Res. 2014;74(2):598–608. | ||
Morales MM, Capella MAM, Sanches MV, Lopes AG, Guggino WB. Modulation of the mdr-1b gene in the kidney of rats subjected to dehydration or a high-salt diet. Pflugers Arch. 2000;439(3):356–362. | ||
Iqbal M, Gibb W, Matthews SG. Corticosteroid regulation of P-glycoprotein in the developing blood-brain barrier. Endocrinology. 2011;152(3):1067–1079. | ||
McShane LM, Altman DG, Sauerbrei W, et al. REporting recommendations for tumour MARKer prognostic studies (REMARK). Br J Cancer. 2005;93(4):387–391. | ||
Vieira-Monteiro Hde A, Freitas-Alves DR, Sobral-Leite M, et al. Prognostic evaluation of VEGFA genotypes and haplotypes in a cohort of Brazilian women with non metastatic breast cancer. Cancer Biol Ther. 2016;17(6):674–683. | ||
Daugherty SL, Powers JD, Magid DJ, et al. Incidence and prognosis of resistant hypertension in hypertensive patients. Circulation. 2012; 125(13):1635–1642. | ||
Ellis IO, Schnitt SJ, Sastre-Garau X, et al. Tumors of the breast. In: Tavassoéli F, Devilee P, editors. Pathology and Genetics of Tumours of the Breast and Female Genital Organs. 3rd ed. Lyon: IARC Press; 2003:9–109. | ||
Elston CW, Ellis IO. Pathological prognostic factors in breast cancer. I. The value of histological grade in breast cancer: experience from a large study with long-term follow-up. Histopathology. 1991;19:403–410. | ||
Huober J, von Minckwitz G, Denkert C, et al. Effect of neoadjuvant anthracycline-taxane-based chemotherapy in different biological breast cancer phenotypes: overall results from the GeparTrio study. Breast Cancer Res Treat. 2010;124(1):133–140. | ||
Mechetner E, Kyshtoobayeva A, Zonis S, et al. Levels of multidrug resistance (MDR1) P-glycoprotein expression by human breast cancer correlate with in vitro resistance to taxol and doxorubicin. Clin Cancer Res. 1998;4(2):389–398. | ||
Bosco DB, Kenworthy R, Zorio DAR, Sang Q-XA. Human mesenchymal stem cells are resistant to Paclitaxel by adopting a non-proliferative fibroblastic state. PLoS One. 2015;10(6):e0128511. | ||
Yang Y, Qiu J-G, Li Y, et al. Targeting ABCB1-mediated tumor multidrug resistance by CRISPR/Cas9-based genome editing. Am J Transl Res. 2016;8(9):3986–3994. | ||
Hung TH, Li YH, Tseng CP, et al. Knockdown of c-MET induced apoptosis in ABCB1-overexpressed multidrug-resistance cancer cell lines. Cancer Gene Ther. 2015;22(5):262–270. | ||
Stebbins MJ, Wilson HK, Canfield SG, Qian T, Palecek SP, Shusta EV. Differentiation and characterization of human pluripotent stem cell-derived brain microvascular endothelial cells. Methods. 2016;101:93–102. | ||
Thiebaut F, Tsuruo T, Hamada H, Gottesman MM, Pastan I, Willingham MC. Cellular localization of the multidrug-resistance gene product P-glycoprotein in normal human tissues. Proc Natl Acad Sci U S A. 1987;84(21):7735–7738. | ||
Thiebaut F, Tsuruo T, Hamada H, Gottesman MM, Pastan I, Willingham MC. Immunohistochemical localization in normal tissues of different epitopes in the multidrug transport protein P170: evidence for localization in brain capillaries and crossreactivity of one antibody with a muscle protein. J Histochem Cytochem. 1989;37(2):159–164. | ||
van Kalken CK, van der Valk P, Hadisaputro MM, et al. Differentiation dependent expression of P-glycoprotein in the normal and neoplastic human kidney. Ann Oncol. 1991;2:55–62. | ||
Sugawara I, Kataoka I, Morishita Y, et al. Tissue distribution of P-glycoprotein encoded by a multidrug-resistant gene as revealed by a monoclonal antibody, MRK 16. Cancer Res. 1988;48(7):1926–1929. | ||
Smit JJ, Schinkel AH, Mol CA, et al. Tissue distribution of the human MDR3 P-glycoprotein. Lab Invest. 1994;71(5):638–649. | ||
Scheffer GL, Kool M, Heijn M, et al. Specific detection of multidrug resistance proteins MRP1, MRP2, MRP3, MRP5, and MDR3 P-glycoprotein with a panel of monoclonal antibodies. Cancer Res. 2000; 60(18):5269–5277. | ||
Kirkegaard T, Edwards J, Tovey S, et al. Observer variation in immunohistochemical analysis of protein expression, time for a change? Histopathology. 2006;48(7):787–794. | ||
Surowiak P, Materna V, Matkowski R, et al. Relationship between the expression of cyclooxygenase 2 and MDR1/P-glycoprotein in invasive breast cancers and their prognostic significance. Breast Cancer Res. 2005;7(5):R862–R870. | ||
Goswami CP, Nakshatri H. PROGgeneV2: enhancements on the existing database. BMC Cancer. 2014;14:970. | ||
Szász AM, Lánczky A, Nagy Á, et al. Cross-validation of survival associated biomarkers in gastric cancer using transcriptomic data of 1,065 patients. Oncotarget. 2016;7(31):49322–49333. | ||
Monteiro Hde AV, Goulart-Citrangulo SMT, Leite MS, Giacomin LC, Vianna-Jorge R. Influência de Variáveis Clinicopatológicas sobre a Eficácia da Quimioterapia Neoadjuvante do Câncer de Mama [Influence of Clinicopathological Variables on the Efficacy of Neoadjuvant]. Rev Bras Cancerol. 2013;59(3):369–377. Portuguese. | ||
Vieira-Monteiro Hde A, Freitas-Alves DR, Sobral-Leite M, et al. Prognostic evaluation of VEGFA genotypes and haplotypes in a cohort of Brazilian women with non metastatic breast cancer. Cancer Biol Ther. 2016;17(6):674–683. | ||
Skaland I, Nordhus M, Gudlaugsson E, et al. Evaluation of 5 different labeled polymer immunohistochemical detection systems. Appl Immunohistochem Mol Morphol. 2010;18(1):90–96. | ||
Charpin C, Vielh P, Duffaud F, et al. Quantitative immunocytochemical assays of P-glycoprotein in breast carcinomas: correlation to messenger RNA expression and to immunohistochemical prognostic indicators. J Natl Cancer Inst. 1994;86(20):1539–1545. | ||
Linn SC, Giaccone G, van Diest PJ, et al. Prognostic relevance of P-glycoprotein expression in breast cancer. Ann Oncol. 1995;6(7):679–685. | ||
Seymour L, Bezwoda WR, Dansey RD. P-glycoprotein immunostaining correlates with ER and with high Ki67 expression but fails to predict anthracycline resistance in patients with advanced breast cancer. Breast Cancer Res Treat. 1995;36(1):61–69. | ||
Charpin C, Bouvier C, Garcia S, et al. Automated and quantitative immunocytochemical assays of Nm23/NDPK protein in breast carcinomas. Int J Cancer. 1997;74(4):416–420. | ||
Charpin C, Garcia S, Bouvier C, et al. Automated and quantitative immunocytochemical assays of Bcl-2 protein in breast carcinomas. Br J Cancer. 1997;76(3):340–346. | ||
Larkin A, O’Driscoll L, Kennedy S, et al. Investigation of MRP-1 protein and MDR-1 P-glycoprotein expression in invasive breast cancer: a prognostic study. Int J Cancer. 2004;112(2):286–294. | ||
Kuroda H, Ishida F, Nakai M, Ohnisi K, Itoyama S. Basal cytokeratin expression in relation to biological factors in breast cancer. Hum Pathol. 2008;39(12):1744–1750. | ||
Pavelic ZP, Sever Z, Fontaine RN, et al. Detection of P-glycoprotein with JSB-1 monoclonal antibody in B-5 fixed and paraffin-embedded cell lines and tissues. Sel Cancer Ther. 1991;7(2):49–58. | ||
Scala S, Saeki T, Lynch A, Salomon D, Merino MJ, Bates SE. Coexpression of TGF alpha, epidermal growth factor receptor, and P-glycoprotein in normal and benign diseased breast tissues. Diagn Mol Pathol. 1995;4(2):136–142. | ||
Zhu Z, Wang B, Bi J, et al. Cytoplasmic HuR expression correlates with P-gp, HER-2 positivity, and poor outcome in breast cancer. Tumour Biol. 2013;34(4):2299–2308. | ||
Doxani C, Voulgarelis M, Zintzaras E. MDR1 mRNA expression and MDR1 gene variants as predictors of response to chemotherapy in patients with acute myeloid leukaemia: a meta-analysis. Biomarkers. 2013; 18(5):425–435. | ||
Sauer G, Kafka A, Grundmann R, Kreienberg R, Zeillinger R, Deissler H. Basal expression of the multidrug resistance gene 1 (MDR-1) is associated with the TT genotype at the polymorphic site C3435T in mammary and ovarian carcinoma cell lines. Cancer Lett. 2002;185(1):79–85. | ||
Taheri M, Mahjoubi F, Omranipour R. Effect of MDR1 polymorphism on multidrug resistance expression in breast cancer patients. Genet Mol Res. 2010;9(1):34–40. | ||
Lopes-Rodrigues V, Seca H, Sousa D, Sousa E, Lima RT, Vasconcelos MH. The network of P-glycoprotein and microRNAs interactions. Int J Cancer. 2014;135(2):253–263. | ||
De La Torre M, Larsson R, Nygren P, Lindgren A, Bergh J. Expression of the multidrug-resistance gene product in untreated human breast cancer and its relationship to prognostic markers. Acta Oncol. 1994;33(7):773–777. | ||
Lu LS, Chen L, Ding WX, Li K, Wu JJ. Elevated expression of both MDR1 and MMP-2 genes in metastasized lymph node of invasive ductal breast cancer. Eur Rev Med Pharmacol Sci. 2012;16(15):2037–2043. | ||
Valente RC, Capella LS, Nascimento CR, et al. ABCB1 (P-glycoprotein) but not ABCC1 (MRP1) is downregulated in peripheral blood mononuclear cells of spontaneously hypertensive rats. Pflugers Arch. 2008;456(2):359–368. | ||
Kang HJ, Song IS, Lee SS, Yoo MA, Shin JG. Effects of dietary salt on the expression of drug transporters, cytochrome P4503a, and nuclear receptors in rats. Xenobiotica. 2008;38:147–155. | ||
Sasano H, Frost AR, Saitoh R, et al. Localization of mineralocorticoid receptor and 11 beta-hydroxysteroid dehydrogenase type II in human breast and its disorders. Anticancer Res. 1997;17(3C):2001–2007. | ||
Tahmasebi M, Barker S, Puddefoot JR, Vinson GP. Localisation of renin-angiotensin system (RAS) components in breast. Br J Cancer. 2006;95(1):67–74. | ||
Mizutani T, Masuda M, Nakai E, et al. Genuine functions of P-glycoprotein (ABCB1). Curr Drug Metab. 2008;9(2):167–174. | ||
Leidy J, Khan A, Kandil D. Basal-like breast cancer: update on clinicopathologic, immunohistochemical, and molecular features. Arch Pathol Lab Med. 2014;138(1):37–43. | ||
van de Vijver MJ, He YD, van’t Veer LJ, et al. A gene-expression signature as a predictor of survival in breast cancer. N Engl J Med. 2002;347(25):1999–2009. | ||
Schmidt M, Böhm D, von Törne C, et al. The humoral immune system has a key prognostic impact in node-negative breast cancer. Cancer Res. 2008;68(13):5405–5413. | ||
Ivshina A V, George J, Senko O, et al. Genetic reclassification of histologic grade delineates new clinical subtypes of breast cancer. Cancer Res. 2006;66(21):10292–10301. | ||
Buffa FM, Camps C, Winchester L, et al. microRNA-associated progression pathways and potential therapeutic targets identified by integrated mRNA and microRNA expression profiling in breast cancer. Cancer Res. 2011;71(17):5635–5645. | ||
Symmans WF, Hatzis C, Sotiriou C, et al. Genomic index of sensitivity to endocrine therapy for breast cancer. J Clin Oncol. 2010;28(27):4111–4119. | ||
Wang Y, Klijn JGM, Zhang Y, et al. Gene-expression profiles to predict distant metastasis of lymph-node-negative primary breast cancer. Lancet. 2005;365(9460):671–679. | ||
Miller LD, Smeds J, George J, et al. An expression signature for p53 status in human breast cancer predicts mutation status, transcriptional effects, and patient survival. Proc Natl Acad Sci U S A. 2005;102(38):13550–13555. | ||
Sabatier R, Finetti P, Cervera N, et al. A gene expression signature identifies two prognostic subgroups of basal breast cancer. Breast Cancer Res Treat. 2011;126(2):407–420. | ||
Center for Computational Biology and Bioinformatics, Indiana University–Purdue University Indianapolis. PROGgeneV2 - Pan Cancer Prognostics Database. Available from: http://watson.compbio.iupui.edu/chirayu/proggene/database/?url=proggene. Accessed May 15, 2017. |
 © 2017 The Author(s). This work is published and licensed by Dove Medical Press Limited. The full terms of this license are available at https://www.dovepress.com/terms.php and incorporate the Creative Commons Attribution - Non Commercial (unported, v3.0) License.
By accessing the work you hereby accept the Terms. Non-commercial uses of the work are permitted without any further permission from Dove Medical Press Limited, provided the work is properly attributed. For permission for commercial use of this work, please see paragraphs 4.2 and 5 of our Terms.
© 2017 The Author(s). This work is published and licensed by Dove Medical Press Limited. The full terms of this license are available at https://www.dovepress.com/terms.php and incorporate the Creative Commons Attribution - Non Commercial (unported, v3.0) License.
By accessing the work you hereby accept the Terms. Non-commercial uses of the work are permitted without any further permission from Dove Medical Press Limited, provided the work is properly attributed. For permission for commercial use of this work, please see paragraphs 4.2 and 5 of our Terms.