Back to Journals » Cancer Management and Research » Volume 11
Loss of Cirbp expression is correlated with the malignant progression and poor prognosis in nasopharyngeal carcinoma
Authors Lin TY, Chen Y , Jia JS, Zhou C, Lian M, Wen YT, Li XY, Chen HW, Lin XL, Zhang XL , Xiao SJ , Sun Y, Xiao D
Received 7 April 2019
Accepted for publication 18 June 2019
Published 24 July 2019 Volume 2019:11 Pages 6959—6969
DOI https://doi.org/10.2147/CMAR.S211389
Checked for plagiarism Yes
Review by Single anonymous peer review
Peer reviewer comments 2
Editor who approved publication: Professor Yong Teng
Tao-Yan Lin,1,* Yan Chen,1,* Jun-Shuang Jia,1,* Chen Zhou,2 Mei Lian,3 Yue-Ting Wen,1 Xiao-Yan Li,3 Heng-Wei Chen,3 Xiao-Lin Lin,1 Xiao-Ling Zhang,4 Sheng-Jun Xiao,2 Yan Sun,5 Dong Xiao1,3
1Guangdong Provincial Key Laboratory of Cancer Immunotherapy Research and Guangzhou Key Laboratory of Tumor Immunology Research, Cancer Research Institute, Southern Medical University, Guangzhou 510515, People’s Republic of China; 2Department of Pathology, The Second Affiliated Hospital, Guilin Medical University, Guilin 541199, People’s Republic of China; 3Institute of Comparative Medicine and Laboratory Animal Center, Southern Medical University, Guangzhou 510515, People’s Republic of China; 4Department of Physiology, Faculty of Basic Medical Sciences, Guilin Medical University, Guilin 541004, People’s Republic of China; 5Zhongshan School of Medicine, Sun Yat-Sen University, Guangzhou 510080, People’s Republic of China
*These authors contributed equally to this work
Purpose: The correlation of cold-inducible RNA-binding protein (Cirbp) expression with clinicopathological features including patient prognosis in nasopharyngeal carcinoma (NPC) was investigated.
Methods: The expression of Cirbp in NPC cell lines and tissue specimens was examined by qRT-PCR or immunohistochemistry (IHC).
Results: Immunohistochemistry (IHC) results showed that high Cirbp expression was detected in 61 of 61 non-cancerous nasopharyngeal squamous epithelial biopsies, whereas the significantly reduced expression of Cirbp was observed in NPC specimens. In addition, IHC assay for Cirbp protein illustrated that the cells of 177 NPC samples and nasopharyngeal squamous epithlial cells displayed strong signals in nuclei and faint signals in cytoplasm, whereas Cirbp protein is mainly detected in the cell’s cytoplasm in many other cancers. More importantly, TNM classification displayed that the low expression of Cirbp was more frequently observed in T3-T4, N2-N3, M1 and III-IV NPC biopsies, and undifferentiated carcinoma (UDC) than T1-T2, N0-N1, M0 and I-II tumors, and differentiated nonkeratinizing carcinoma (DNKC), suggesting that Cirbp loss is a key molecular event in advanced cases of NPC. Kaplan–Meier survival analysis indicated that NPC patients showing lower Cirbp expression had a significantly shorter overall survival time than those with high Cirbp expression. Multivariate analysis suggested that the level of Cirbp expression was an independent prognostic indicator for NPC survival. Finally, we revealed a significant positive association between Cirbp expression and E-cadherin, and a notable negative correlation between Cirbp expression and Ki67 labeling index in NPC biopsies.
Conclusion: Collectively, these findings demonstrate that loss of Cirbp expression is correlated with malignant progression and poor prognosis in NPC.
Keywords: cirbp, nasopharyngeal carcinoma; NPC, TNM stage, prognosis, E-cadherin, Ki67
Introduction
Human nasopharyngeal carcinoma (NPC) is a squamous cell carcinoma originated from the human nasopharyngeal mucosal epithelium.1,2 NPC is common in southern China (mainly in Guangdong province) with an incidence rate of about 20 per 100,000 people each year.1,2 Unfortunately, the majority of NPC patients tend to present with more advanced disease when first diagnosed because of its deep location and vague symptoms.1,2 As such, the local invasion and early distant metastases are often present at initial diagnosis, thereby leading to a poor prognosis in the patients with NPC.1,2 Thus, the better understanding of the cellular and molecular mechanisms involved in the NPC pathogenesis is essential for early diagnosis, prognostic prediction and the development of novel therapeutic strategies for NPC patients.
Cold-inducible RNA-binding protein (Cirbp, also known as A18 hnRNP or CIRP) is a member of the cold shock protein family and a general stress-response protein that is induced by varieties of cellular stresses, such as UV irradiation, hypoxia, heat shock and cold treatment.3-7 Cirbp is ubiquitously expressed in wide variety of tissues and cells of human and mice, including brain, lung, heart, kidney, retina, testis, liver, ovary, lymphocytes and endometrium, while Cirbp is mainly localized in the nucleus, but can translocate from the nucleus into the cytosol under stress conditions.3,4,6–8 Cirbp exerts its function either through its RNA-binding activity by binding its target genes intracellularly or acts as a secreted damage-associated molecular pattern triggering inflammatory response extracellularly.3,4,6–8 Through the regulation of its targets, Cirbp has been implicated in diverse physiological and pathological processes, including cell proliferation and differentiation, cell senescence, cell survival and apoptosis, oxidative stress, DNA damage and repair, immune and inflammatory responses, telomere maintenance, circadian rhythm, spermatogenesis, tumor formation and progression, etc.3,4,6–8 Recent study reported that PARP-1–dependent recruitment of Cirbp promotes double-strand break repair and genome stability.3 In summary, Cirbp is a highly conserved multifunctional protein in vertebrates and expressed in various cells and participates in various physiological and pathological processes in vivo. A better understanding of the detailed mechanisms of Cirbp upregulated expression and translocation still needs to be further studied.
Cirbp plays a divergent role in various cancers, depending on different cell context. Evidences from these studies in rectal carcinoma, endometrial carcinoma and ovarian tumor showed that Cirbp functions as a tumor suppressor, whereas Cirbp exerts its pro-tumorigenic influences on melanoma, breast cancer, colorectal cancer, prostate cancer, central-nervous-system-related tumor, liver-pancreas carcinomas, skin squamous cell carcinoma, bladder cancer and pituitary corticotroph adenoma,3–8 suggesting that Cirbp has context-dependent oncogenic and tumor suppressor functions in cancer progression. However, the involvement of Cirbp in NPC progression has not yet been investigated.
To dissect the functions of Cirbp in NPC pathogenesis, the purpose of this study is to investigate the expression profile of Cirbp in human NPC clinical tissue biopsies, and subsequently analyze the potential associations of this protein expression levels with clinicopathological features, prognosis and overall survival in NPC patients.
Materials and methods
Cell lines and cell culture
Human NPC cell lines (ie, CNE1, CNE2, SUNE1, HONE1, HONE1-EBV, S18, S26 and 5-8F) and human immortalized nasopharyngeal epithelial cell line (ie, NP69 cells) were kindly provided from Prof. Qiao Tao (Chinese University of Hong Kong, Hong Kong, China), Prof. S.-W. Tsao (University of Hong Kong), Prof. Yixin Zeng (Sun Yat-Sen University, Guangzhou, China) and Prof. Musheng Zeng (Sun Yat-sen University, Guangzhou, China). NPC cell lines and NP69 cell line were approved by the Institutional Review Board of Southern Medical University. NPC cell lines were cultured in RPMI 1640 medium supplemented with 10% fetal bovine serum (FBS) in a humidified incubator with 5% CO2 at 37°C, while NP69 cell line was maintained in keratinocyte/serum-free medium (Invitrogen).
Patients and tumor tissues
14 fresh NPC tissue specimens and 14 fresh non-cancerous nasopharyngeal epithelial tissues (NP) (ie, chronic nasopharyngitis tissues) (for qRT-PCR) were collected from the Department of Otorhinolaryngology, Head and Neck Surgery, Zhujiang Hospital, Southern Medical University, Guangzhou, China, while other 24 fresh NPC tissue specimens and 9 fresh non-cancerous nasopharyngeal epithelial tissue samples were collected from the Department of Pathology, The Second Affiliated Hospital, Guilin Medical University, Guilin, China. None of the NPC patients received preoperative radiotherapy or chemotherapy. The clinicopathologic variables and related information of NPC biopsies were collected as previously described.9,10 All fresh samples were frozen immediately in liquid nitrogen. This study was approved by the Ethics Committee of Zhujiang Hospital of Southern Medical University and The Second Affiliated Hospital of Guilin Medical University. Before the use of these clinical samples, informed consent from the patients was obtained.
Furthermore, we used a tissue microarray (TMA) encompassing 61 non-cancerous nasopharyngeal epithelia tissue punches and 177 NPC tissue punches from formalin-fixed paraffin-embedded tumor samples collected from patients diagnosed with primary NPC between 2005 and 2010 at the Department of Pathology, the Second Affiliated Hospital of Guilin Medical University in Guilin, China. The application of tissue microarray was approved by the Ethics Committee of the Second Affiliated Hospital of Guilin Medical University. Histopathologic data were obtained from the pathology reports, and raw patient survival data were obtained from the patient’s attending physician. Retrieval of tissue and clinical data was performed according to the regulations of the institutional review boards (IRB) of the Second Affiliated Hospital of Guilin Medical University and data safety laws with specific regard to ethical standards and patient confidentiality. The follow-up time was 60 months, and the mean age of the patients at diagnosis was 63 years (range 18–78 years).
RNA isolation and quantitative real-time PCR (qRT-PCR)
RNA isolation and qRT-PCR were previously well described.11–16 Briefly, total RNA was extracted from cells or fresh tissue specimens using Trizol Reagent (TaKaRa, Dalian, China) according to the manufacturer’s instruction. Then, mRNA was reversely transcribed to cDNA using the PrimeScript RT reagent Kit (TaKaRa). To evaluate the mRNA levels of a number of genes, qRT-PCR was performed on LightCycler® 96 qRT-PCR system (Roche, Switzerland) using SYBR Premix Ex Taq (Takara). GAPDH was used as the internal control. GAPDH forward primer: 5′-ACCCAGAAGACTGTGGATGG-3′, and GAPDH reverse primer: 5′-TCTAGACGGCAGGTCAGGTC-3′; Cirbp forward primer: 5′-AGGGCTGA GTTTTGACACCAA-3′, and Cirbp reverse primer: 5′-ACAAACCCAAATCCCCGAGAT-3′; E-cadherin forward primer: 5′- ATTTTTCCCTCGACACCCGAT-3′, and E-cadherin reverse primer: 5′-TCCCAGGCGTAGACCAAGA-3′; Ki67 forward primer: 5′- TATCAAAAGGAGCGGGGTCG-3′, and Ki67 reverse primer: 5′- TTTCATACCTTCCAAACAAGCAG-3′. All samples were normalized to internal controls and fold changes were calculated based on relative quantification (2−△△Ct).
Immunohistochemistry (IHC)
Histological analysis and immunohistochemical staining were performed as described previously.11–14,17,18 Briefly, for immunohistochemical staining, 4-µm sections of the TMA blocks were deparaffinized, rehydrated. Then the paraffin-embedded sections were subjected to high pressure for 3 mins for antigenic retrieval in sodium citrate buffer (10 mM Sodium Citrate, 0.05% Tween 20, pH 6.0) for 3 mins. After antigenic retrieval, the sections were blocked with 3% H2O2 for 10 mins, and subsequently incubated overnight at 4°C with the following primary antibodies: Cirbp (Proteintech, Cat No. 10,209–2-AP), E-cadherin (BD, Cat No. 610,182) and Ki67 (Abcam, Cat No. ab16667). The slides were then stained with a goat anti-rabbit secondary antibody (UltraSensitiveTM SP IHC Kit, Maxim biotechnologies, Fuzhou, China; Cat No. KIT-0105R) at 37°C for 20 mins. Finally, the sections were stained with diaminobenzidine (DAB) and counterstained with hematoxylin. As Cirbp was expressed in the nucleus of almost all tumor cells and nasopharyngeal epithelial cells, only staining intensity was evaluated for IHC score. Staining intensity is classified as 0 (negative); 1 (weak); 2 (moderate); and 3 (strong). The scores 0 and 1 are defined as low, and the scores 2 and 3 are defined as high. Histopathology and immunohistochemistry were evaluated independently by two pathologists without knowledge of the clinicopathological results of the patients.
Statistical analyses
Statistical analyses were performed using the SPSS 20.0 software package. The χ2 test was used to analyze the association between clinicopathological characteristics and Cirbp expression. Overall survival (OS) was measured from the onset of treatment to the date of death or the survival status at the last date of follow-up. OS probabilities were estimated by the Kaplan–Meier method and the significance of differences was assessed by the log-rank test. Two-tailed Student’s t-test was used for comparisons of two independent groups. The data were presented as mean ± SEM. Values are statistically significant at *P<0.05, **P<0.01 and #P<0.001.
Results
Cirbp mRNA is expressed at low levels in NPC cell lines and tissue specimens
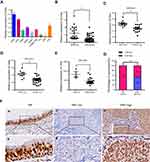
To evaluate the potential functions of Cirbp in NPC progression, qRT-PCR assay was performed to quantify the transcript abundance of Cirbp in eight NPC cell lines, 38 freshly collected NPC tissues and 23 freshly collected non-cancerous nasopharyngeal biopsies (NP tissues). Our results revealed that the expression of Cirbp mRNA transcripts was markedly down-regulated in NPC cell lines, such as CNE1, CNE2, SUNE1, HONE1, HONE1-EBV, S18, S26 and 5-8F (Figure 1A), compared with NP69 cells. Compared with NP tissues, NPC tissue biopsies displayed the decreased expression levels of Cirbp mRNA (P<0.001) (Figure 1B). Additionally, three GEO datasets revealed that the down-regulated expression of Cirbp was observed in NPC tissues as compared with NP tissues (Figure 1C,D,E). Collectively, the qRT-PCR analysis and GEO datasets clearly showed that the reduced Cirbp expression was frequently detected in NPC cell lines and NPC clinical tissue biopsies.
Immunohistochemical analysis of Cirbp protein expression in NPC and NP tissues
Next, we examined the expression levels and subcellular localization of Cirbp protein in 177 paraffin-embedded, archived NPC biopsies and 61 paraffin-embedded, archived non-cancerous NP samples using immunohistochemistry (IHC) staining. IHC results revealed that high expression of Cirbp was detected in 61 of 61 non-cancerous NP squamous epithelial cells (100%)(Figure 1F,G and Table 1). In contrast, among 177 NPC samples, 95 samples (53.7%) exhibited low or no expression of Cirbp and 82 samples (46.3%) showed high Cirbp staining (Figure 1F,G and Table 1). Therefore, the results from IHC staining demonstrate that Cirbp downregulation more frequently occurred in NPC tissue biopsies than in their non-cancerous counterparts.
 |
Table 1 Expression of Cirbp in 61 non-cancerous nasopharyngeal (NP) epithelial tissues and 177 nasopharyngeal carcinoma (NPC) tissues |
Furthermore, IHC assay for Cirbp protein subcellular localization illustrated that the cells of 177 NPC samples and nasopharyngeal squamous epithelial cells displayed strong signals in nuclei and faint signals in cytoplasm (Figures 1F and 2A), whereas Cirbp protein is mainly detected in the cell’s cytoplasm in many other cancers.
Cirbp downregulation in NPC specimens was associated with aggressive and/or poor prognostic phenotypes of NPC
The relationship between Cirbp expression and a number of clinicopathologic characteristics of the patients with NPC was summarized in Table 2. No significant association was identified between Cirbp expression and the age (P=0.781) and sex (P=0.883) of 177 NPC cases (Table 2). However, we found that Cirbp expression level was correlated inversely with the tumor size (T classification)(P<0.001), lymph node invasion (N classification) (P<0.001), distant metastasis (M classification)(P=0.001), clinical stage (I–II versus III–IV) (P<0.001) and histological subtype (P=0.002) in 177 NPC patients (Figure 2A,B and Table 2). Briefly, low expression of Cirbp was more frequently detected in T3-T4, N2-N3, M1 and III-IV NPC biopsies, and undifferentiated carcinoma (UDC) than T1-T2, N0-N1, M0 and I-II tumors, and differentiated nonkeratinizing carcinoma (DNKC) (Figure 2A,B and Table 2), indicating that Cirbp loss is a key molecular event in advanced cases of NPC.
 |
Table 2 Association between the clinicopathological features and Cirbp expression in 177 nasopharyngeal carcinoma (NPC) tissue specimens |
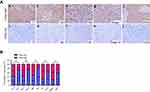
Moreover, given that Cirbp downregulation was associated with advanced NPC, we further evaluated the prognostic value of Cirbp expression for NPC patients. A negative correlation between the level of Cirbp protein expression and the overall survival of NPC patients was identified based on Kaplan–Meier analysis of the log-rank test (Figure 3F). Kaplan–Meier survival analysis indicated that the patients with lower levels of Cirbp expression had poorer overall survival than those with higher levels of Cirbp expression (P=0.009; Figure 3F). Kaplan–Meier survival analysis indicated that patients who had high Cirbp expression levels had a better outcome (P=0.0089; Figure 3F). Additionally, univariate analyses revealed that histological type, T, N and M classification, and clinical stage were associated significantly with the overall survival of NPC patients (P=0.046, P=0.020, P=0.018, P<0.001 and P=0.024, respectively; Figure 3A,B,C, Figure 3A,D, Figure 3A,E and Table 3). Multivariate analysis was performed to identify independent prognostic factors for the patients with NPC. Multivariate Cox regression analysis indicated that age (P=0.015), histological type (P=0.000), lymph node metastasis (P=0.008), distant metastasis (P=0.000), clinical stage (P=0.008) and Cirbp expression (P=0.000) were independent prognostic factors for NPC patients (Table 3). Therefore, Cirbp downregulation in advanced cases of NPC was markedly associated with the poor prognosis of patients with NPC, whereas Cirbp expression was an independent prognostic factor for NPC patients.
 |
Table 3 Univariate and multivariate analyses of the overall survival of 177 patients with nasopharyngeal carcinoma (NPC) |
 |
Table 4 Correlation between Cirbp and E-cadherin & Ki6 in 177 nasopharyngeal carcinoma (NPC) tissue specimens |
Association between Cirbp expression and e-cadherin or Ki67 expression in clinical NPC specimens
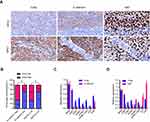
We further identified the association between the expression of EMT-related marker (ie, E-cadherin) and Cirbp in 177 cases of human primary NPC tissues. Representative pictures of immunohistochemical staining in NPC tissues are shown in Figure 4A. Statistical analyses revealed a significant positive association between E-cadherin and Cirbp expression in NPC biopsies (Figure 4B and Table 4). We further analyzed the expression levels of Cirbp and E-cadherin in NP69 and NPC cell lines, and qRT-PCR analysis revealed that Cirbp and E-cadherin were markedly downregulated at mRNA levels when compared with NP69 (Figure 4C). Summarily, Cirbp might be also associated with reduced EMT phenotype in human NPC tissues.
We further determined the association between the proliferation index Ki67 and Cirbp expression in 177 cases of human primary NPC tissues. Representative pictures of immunohistochemical staining in NPC tissues are shown in Figure 3A. Chi-square test showed a significant negative correlation between Cirbp expression and Ki67 labeling index in NPC (P<0.001) (Figure 4B and Table 4). qRT-PCR analysis revealed that Cirbp and Ki67 were markedly downregulated at mRNA levels when compared with NP69 (Figure 4D). Briefly, NPC samples which had lower level of Cirbp expression also had a higher Ki67 labeling index, whereas NPC samples which had lower level of Cirbp expression had a higher Ki67 labeling index (Figure 4B and Table 4).
Discussion
It is well known that NPC is a unique malignant head and neck cancer with a remarkably distinctive global ethnic and geographic distribution among the world.1,2 The majority of NPC-related deaths are attributed to distant metastasis rather than the primary tumor.1,2 However, the molecular mechanisms of NPC malignant progression are not yet well understood.
Accumulated evidence demonstrates the tumor suppressor or oncogenic functions of Cirbp in the progression of various cancers.3–8 In this study, we observed the significantly reduced expression of Cirbp in NPC cell lines and tissue specimens. Similarly, the decreased expression of Cirbp was observed in endometrial19 and ovarian cancer.20 Cirbp’s initial functions as a tumor suppressor were established during hypothermic stress and DNA damage.21,22 The previous studies revealed that Cirbp may function as a tumor suppressor via suppressing cell proliferation, potentially via its function in the DNA damage response.19,20,23,24 In contrast, Cirbp with capability of promoting cell proliferation is upregulated in a subgroup of patients with solid cancers, including melanoma, prostate, breast, colon, central-nervous-system-related, liver and bladder cancers, oral squamous cell carcinoma and pituitary adenoma, and functions as a proto-oncogene in the aforementioned cancers.3–8,25–28 More importantly, this study revealed a tendency toward lower Cirbp expression in samples from patients of clinical stages III–IV, T3-T4, N2-N3, M1 or UDC compared to those of I–II, T1–T2, N0–N1, M1 or DNKC, respectively. Furthermore, our IHC results showed that the level of Cirbp expression inversely correlated with the proliferation marker Ki67 in human NPC clinical tissue specimens, while Cirbp expression level was correlated inversely with the tumor size (T classification) in 177 NPC patients. Additionally, the high levels of Cirbp expression in human breast cancer,29 liver cancer,30,31 colon cancer,32 pituitary adenoma,27,33 oral squamous cell carcinoma28 are associated with a short survival rate or poor prognosis,4,6,7 whereas we found that the low levels of Cirbp expression in NPC are associated with a short survival rate. These findings from NPC cell lines and tissue specimens strongly suggested that Cirbp might function as a tumor suppressor in the pathogenesis of NPC, which remains to be fully investigated.
Cirbp is mainly localized in the nucleus, but under various stresses (ie, hypoxia and UV radiation). Cirbp protein can translocate from the nucleus to the cytoplasm, and then regulate mRNA stability by binding to the 3ʹ-UTR of stress-responsive transcripts.3,4,6,7,8 Cirbp is downregulated in endometrial and ovarian cancers, and upregulated in solid cancers including human colon cancer, liver cancer, prostate cancer, central-nervous-system-related cancer and squamous cell carcinoma, in which Cirbp protein is mainly detected in the cytoplasm of tumor cells, indicating that Cirbp cytoplasmic translocation may have biological significances in human tumor progression.6,7,19,20,25,34–36 The cytoplasmic localization of Cirbp protein in cancers is consistent with the fact that most solid tumors develop hypoxic regions, and Cirbp protein translocates to the cytosol in response to hypoxia.6,7,19,20,25,34,35,37,38 In contrast, the subcellular localization of Cirbp protein was mainly found in the nuclei of tumor cells among 177 NPC samples and nasopharyngeal epithelial cells in 61 non-cancerous NP tissue samples, whereas the faint signals of Cirbp protein were observed in the cytoplasm of NPC tissue cells and nasopharyngeal epithelial cells. More direct evidences and details about Cirbp functions in NPC cell nuclei need to be further revealed.
Multiple high-throughput screening methods revealed that a panel of adhesion molecules, including αE- and β-catenin, C- and E-cadherin and paraxial proto-cadherin, has been identified as the targets of Cirbp in Xenopus.39,40 Our results showed a significant positive association between E-cadherin and Cirbp expression in NPC biopsies. These results suggest broad targets and multiple roles of Cirbp in NPC progression (ie, EMT, invasion and metastasis), which remains to be fully examined.
Conclusion
In summary, the present study demonstrates that the expression levels of Cirbp are significantly decreased in NPC cell lines and tissue specimens, and the loss of Cirbp expression is closely correlated with malignant progression and poor prognosis in NPC. Additionally, further functional studies will be required to fully dissect the roles of Cirbp in NPC progression and underlying mechanisms.
Abbreviation list
Cirbp, cold-inducible RNA-binding protein; DNKC, differentiated non-keratinizing carcinoma; EMT, epithelial-mesenchymal transition; IHC, immunohistochemistry; NPC, nasopharyngeal carcinoma; TMA, tissue microarray; UDC, undifferentiated carcinoma.
Acknowledgments
This work was supported by the National Natural Science Foundation of China (Grant No. 81872209, 81672689, 81372896 and 81172587, to D Xiao; Grant No. 81600086 and 81770100, to Y Sun; Grant No. 81600488 and 81870602, to XL Lin; Grant No. 81702778, to JS Jia; Grant No. 81560441 and 81760491, to SJ Xiao), the Science and Technology Planning Project of Guangdong Province of China (Grant No. 2017A030303018, to JS Jia) and the Medical Scientific Research Foundation of Guangdong Province of China (Grant No. A2017420, to JS Jia).
Author contributions
D Xiao, Y Sun and SJ Xiao conceived and designed the study; TY Lin, SJ Xiao, Y Chen, JS Jia, C Zhou, M Lian, YT Wen, XY Li and HW Chen performed the experiments; D Xiao, SJ Xiao and TY Lin analyzed the data; XL Lin and XL Zhang contributed essential reagents or tools; D Xiao, TY Lin and SJ Xiao wrote the paper. All authors contributed to data analysis, drafting and revising the article, gave final approval of the version to be published, and agree to be accountable for all aspects of the work.
Disclosure
The authors report no conflicts of interest in this work.
References
1. Chua MLK, Wee JTS, Hui EP, Chan ATC. Nasopharyngeal carcinoma. Lancet (London, England). 2016;387:1012–1024. doi:10.1016/S0140-6736(15)00055-0
2. Torre LA, Bray F, Siegel RL, Ferlay J, Lortet-Tieulent J, Jemal A. Global cancer statistics, 2012. CA Cancer J Clin. 2015;65:87–108. doi:10.3322/caac.21262
3. Chen JK, Lin WL, Chen Z, Liu HW. Parp-1-dependent recruitment of cold-inducible RNA-binding protein promotes double-strand break repair and genome stability. Proc Natl Acad Sci U S A. 2018;115:E1759–e1768. doi:10.1073/pnas.1713912115
4. Liao Y, Tong L, Tang L. The role of cold-inducible RNA binding protein in cell stress response. Int J Cancer. 2017;141:2164–2173. doi:10.1002/ijc.30833
5. Lu M, Ge Q, Wang G, et al. Cirbp is a novel oncogene in human bladder cancer inducing expression of hif-1alpha. Cell Death Dis. 2018;9:1046. doi:10.1038/s41419-018-1111-y
6. Zhong P, Huang H. Recent progress in the research of cold-inducible RNA-binding protein. Future Sci OA. 2017;3:Fso246. doi:10.4155/fsoa-2017-0077
7. Zhu X, Buhrer C, Wellmann S. Cold-inducible proteins cirp and rbm3, a unique couple with activities far beyond the cold. Cell Mol Life Sci. 2016;73:3839–3859. doi:10.1007/s00018-016-2253-7
8. Lujan DA, Ochoa JL, Hartley RS. Cold-inducible RNA binding protein in cancer and inflammation. Wiley Interdiscip Rev RNA. 2018;9. doi:10.1002/wrna.1462
9. Li X, Zhao Z, Zhang X, et al. Klf4 reduces stemness phenotype, triggers mesenchymal-epithelial transition (met)-like molecular changes, and prevents tumor progression in nasopharygeal carcinoma. Oncotarget. 2017;8:93924–93941. doi:10.18632/oncotarget.21370
10. Wang SC, Lin XL, Wang HY, et al. Hes1 triggers epithelial-mesenchymal transition (emt)-like cellular marker alterations and promotes invasion and metastasis of nasopharyngeal carcinoma by activating the pten/akt pathway. Oncotarget. 2015;6:36713–36730. doi:10.18632/oncotarget.5457
11. Liu YM, Liu W, Jia JS, et al. Abnormalities of hair structure and skin histology derived from crispr/cas9-based knockout of phospholipase c-delta 1 in mice. J Transl Med. 2018;16:141. doi:10.1186/s12967-018-1512-9
12. Lin X, Jia J, Du T, et al. Overexpression of mir-155 in the liver of transgenic mice alters the expression profiling of hepatic genes associated with lipid metabolism. PLoS One. 2015;10:e0118417. doi:10.1371/journal.pone.0118417
13. Lin X, Zhao W, Jia J, et al. Ectopic expression of cripto-1 in transgenic mouse embryos causes hemorrhages, fatal cardiac defects and embryonic lethality. Sci Rep. 2016;6:34501. doi:10.1038/srep34501
14. Xie R, Lin X, Du T, et al. Targeted disruption of mir-17-92 impairs mouse spermatogenesis by activating mtor signaling pathway. Medicine. 2016;95:e2713. doi:10.1097/MD.0000000000004864
15. Li J, Yang S, Yan W, et al. Microrna-19 triggers epithelial-mesenchymal transition of lung cancer cells accompanied by growth inhibition. Lab Invest. 2015;95:1056–1070. doi:10.1038/labinvest.2015.76
16. Wang SC, Lin XL, Li J, et al. Microrna-122 triggers mesenchymal-epithelial transition and suppresses hepatocellular carcinoma cell motility and invasion by targeting rhoa. PLoS One. 2014;9:e101330. doi:10.1371/journal.pone.0101330
17. Rong XX, Wei F, Lin XL, et al. Recognition and killing of cancer stem-like cell population in hepatocellular carcinoma cells by cytokine-induced killer cells via nkg2d-ligands recognition. Oncoimmunology. 2016;5:e1086060. doi:10.1080/2162402X.2015.1086060
18. Sun Y, Xiao D, Li HA, et al. Phenotypic changes of human cells in human-rat liver during partial hepatectomy-induced regeneration. World J Gastroenterol. 2009;15:3611–3620. doi:10.3748/wjg.15.3611
19. Hamid AA, Mandai M, Fujita J, et al. Expression of cold-inducible RNA-binding protein in the normal endometrium, endometrial hyperplasia, and endometrial carcinoma. Int J Gynecol Pathol. 2003;22:240–247. doi:10.1097/01.PGP.0000070851.25718.EC
20. Biade S, Marinucci M, Schick J, et al. Gene expression profiling of human ovarian tumours. Br J Cancer. 2006;95:1092–1100. doi:10.1038/sj.bjc.6603346
21. Nishiyama H, Itoh K, Kaneko Y, Kishishita M, Yoshida O, Fujita J. A glycine-rich RNA-binding protein mediating cold-inducible suppression of mammalian cell growth. J Cell Biol. 1997;137:899–908. doi:10.1083/jcb.137.4.899
22. Sheikh MS, Carrier F, Papathanasiou MA, et al. Identification of several human homologs of hamster DNA damage-inducible transcripts. Cloning and characterization of a novel uv-inducible cdna that codes for a putative RNA-binding protein. J Biol Chem. 1997;272:26720–26726. doi:10.1074/jbc.272.42.26720
23. Lujan DA, Garcia S, Vanderhoof J, et al. Cold-inducible RNA binding protein in mouse mammary gland development. Tissue Cell. 2016;48:577–587. doi:10.1016/j.tice.2016.10.004
24. Peri S, de Cicco RL, Santucci-Pereira J, et al. Defining the genomic signature of the parous breast. BMC Med Genomics. 2012;5:46. doi:10.1186/1755-8794-5-46
25. Artero-Castro A, Callejas FB, Castellvi J, et al. Cold-inducible RNA-binding protein bypasses replicative senescence in primary cells through extracellular signal-regulated kinase 1 and 2 activation. Mol Cell Biol. 2009;29:1855–1868. doi:10.1128/MCB.01386-08
26. Chang ET, Parekh PR, Yang Q, Nguyen DM, Carrier F. Heterogenous ribonucleoprotein a18 (hnrnp a18) promotes tumor growth by increasing protein translation of selected transcripts in cancer cells. Oncotarget. 2016;7:10578–10593. doi:10.18632/oncotarget.7020
27. Jian F, Chen Y, Ning G, et al. Cold inducible rna binding protein upregulation in pituitary corticotroph adenoma induces corticotroph cell proliferation via erk signaling pathway. Oncotarget. 2016;7:
28. Ren WH, Zhang LM, Liu HQ, et al. Protein overexpression of cirp and tlr4 in oral squamous cell carcinoma: an immunohistochemical and clinical correlation analysis. Med Oncol. 2014;31:120. doi:10.1007/s12032-014-0374-0
29. Guo X, Wu Y, Hartley RS. Cold-inducible RNA-binding protein contributes to human antigen r and cyclin e1 deregulation in breast cancer. Mol Carcinog. 2010;49:130–140. doi:10.1002/mc.20582
30. Lee HN, Ahn SM, Jang HH. Cold-inducible RNA-binding protein, cirp, inhibits DNA damage-induced apoptosis by regulating p53. Biochem Biophys Res Commun. 2015;464:916–921. doi:10.1016/j.bbrc.2015.07.066
31. Sakurai T, Yada N, Watanabe T, et al. Cold-inducible RNA-binding protein promotes the development of liver cancer. Cancer Sci. 2015;106:352–358. doi:10.1111/cas.12611
32. Sakurai T, Kashida H, Watanabe T, et al. Stress response protein cirp links inflammation and tumorigenesis in colitis-associated cancer. Cancer Res. 2014;74:6119–6128.
33. Wang M, Zhang H, Heng X, Pang Q, Sun A. Expression of cold-inducible rna-binding protein (cirp) in pituitary adenoma and its relationships with tumor recurrence. Med Sci Monit. 2015;21:1256–1260. doi:10.12659/MSM.893128
34. Liao Y, Feng J, Zhang Y, Tang L, Wu S. The mechanism of cirp in inhibition of keratinocytes growth arrest and apoptosis following low dose uvb radiation. Mol Carcinog. 2017;56:1554–1569. doi:10.1002/mc.22597
35. Park BM, Lee JH, Kim SJ. Expression of cold-inducible RNA-binding protein in normal skin, actinic keratosis and squamous cell carcinoma. Ann Dermatol. 2014;26:256–258. doi:10.5021/ad.2014.26.2.256
36. Yang R, Weber DJ, Carrier F. Post-transcriptional regulation of thioredoxin by the stress inducible heterogenous ribonucleoprotein a18. Nucleic Acids Res. 2006;34:1224–1236. doi:10.1093/nar/gkj519
37. Harris AL. Hypoxia – a key regulatory factor in tumour growth. Nat Rev Cancer. 2002;2:38–47. doi:10.1038/nrc704
38. Yang R, Zhan M, Nalabothula NR, Yang Q, Indig FE, Carrier F. Functional significance for a heterogenous ribonucleoprotein a18 signature rna motif in the 3ʹ-untranslated region of ataxia telangiectasia mutated and rad3-related (atr) transcript. J Biol Chem. 2010;285:8887–8893. doi:10.1074/jbc.M109.013128
39. Morf J, Rey G, Schneider K, et al. Cold-inducible RNA-binding protein modulates circadian gene expression posttranscriptionally. Science. 2012;338:379–383. doi:10.1126/science.1217726
40. Peng Y, Yang PH, Tanner JA, et al. Cold-inducible RNA binding protein is required for the expression of adhesion molecules and embryonic cell movement in xenopus laevis. Biochem Biophys Res Commun. 2006;344:416–424. doi:10.1016/j.bbrc.2006.03.086
 © 2019 The Author(s). This work is published and licensed by Dove Medical Press Limited. The full terms of this license are available at https://www.dovepress.com/terms.php and incorporate the Creative Commons Attribution - Non Commercial (unported, v3.0) License.
By accessing the work you hereby accept the Terms. Non-commercial uses of the work are permitted without any further permission from Dove Medical Press Limited, provided the work is properly attributed. For permission for commercial use of this work, please see paragraphs 4.2 and 5 of our Terms.
© 2019 The Author(s). This work is published and licensed by Dove Medical Press Limited. The full terms of this license are available at https://www.dovepress.com/terms.php and incorporate the Creative Commons Attribution - Non Commercial (unported, v3.0) License.
By accessing the work you hereby accept the Terms. Non-commercial uses of the work are permitted without any further permission from Dove Medical Press Limited, provided the work is properly attributed. For permission for commercial use of this work, please see paragraphs 4.2 and 5 of our Terms.