Back to Journals » OncoTargets and Therapy » Volume 13
Long Noncoding RNAs in Diffuse Large B-Cell Lymphoma: Current Advances and Perspectives
Received 10 March 2020
Accepted for publication 28 April 2020
Published 18 May 2020 Volume 2020:13 Pages 4295—4303
DOI https://doi.org/10.2147/OTT.S253330
Checked for plagiarism Yes
Review by Single anonymous peer review
Peer reviewer comments 2
Editor who approved publication: Dr Sanjay Singh
Xianbo Huang,1 Wenbin Qian,2 Xiujin Ye1
1Department of Hematology, The First Affiliated Hospital, College of Medicine, Zhejiang University, Hangzhou 310003, People’s Republic of China; 2Department of Hematology, The Second Affiliated Hospital, College of Medicine, Zhejiang University, Hangzhou 310003, People’s Republic of China
Correspondence: Xiujin Ye
Tel +86-571-87235875
Fax +86-571-87235560;
Email [email protected]
Wenbin Qian
Tel/Fax +86-571-89713674
Email [email protected]
Abstract: Diffuse large B-cell lymphoma (DLBCL) is a complex and aggressive malignancy originating from B lymphocytes and characterized by extensive clinical, phenotypic and molecular heterogeneity. Although research conducted over the past decades has substantially improved our understanding of DLBCL, its pathogenesis has not yet been fully elucidated. The development of RNA sequencing technology has allowed the identification of numerous long noncoding RNAs (lncRNAs) that exhibit aberrant expression in DLBCL. These lncRNAs play crucial roles in DLBCL development and pathogenesis and are thus good candidates for use as diagnostic biomarkers or therapeutic targets. In this review, we describe the lncRNAs associated with DLBCL, summarize their characteristics and molecular functions, and discuss their relationships with clinical practice.
Keywords: diffuse large B-cell lymphoma, long noncoding RNA, biomarkers
Introduction
Diffuse large B-cell lymphoma (DLBCL), which is a highly aggressive lymphoid malignancy characterized by large B lymphoid cells exhibiting a diffuse growth pattern,1 is the most prevalent subtype of non-Hodgkin lymphoma (NHL), accounting for 25–35% of all NHL cases globally.2 The standard-of-care chemoimmunotherapeutic regimen, which consists of R-CHOP, typically leads to a complete response in 50–70% of patients.3 Unfortunately, 30–40% of patients are either refractory to the current treatment regimen or experience disease relapse after complete response, and thus, these patients exhibit a dismal prognosis.4,5 Therefore, a better understanding of the molecular mechanism, the exploration of new biomarkers for the diagnosis of DLBCL and prognosis prediction, and the discovery of novel therapeutic targets of DLBCL would be helpful for improving the curative effect of the developed treatments and the prognosis of patients with this disease.
The discovery of noncoding RNAs (ncRNAs) in recent decades has revealed new prospects for tumor research. ncRNAs, including long noncoding RNAs (lncRNAs), microRNAs (miRNAs) and circular RNAs, are RNA transcripts of variable length that are not translated into proteins.6 The rapid development of next-generation sequencing and bioinformatic approaches has revealed that these functional RNA molecules are greatly relevant to cancer biology.7 lncRNAs, which are defined as ncRNAs containing more than 200 nucleotides located within intergenic, intronic, or antisense stretches within or overlap with protein-coding genes,8 are one of the most studied types of ncRNAs and perform important functions in the modulation of gene expression at multiple levels. Emerging lines of evidence indicate that lncRNAs are critical regulators and actively participate in the physiological and pathological processes of human cancers, including hematological malignancies.9 To date, the roles and mechanisms of abnormally expressed lncRNAs in DLBCL have not been fully clarified. In this review, the PubMed database was searched for articles written in the English language that include combinations of the Medical Subject Headings terms long noncoding RNA, lncRNA, lymphoma, diffuse large B cell lymphoma and DLBCL. We aim to provide a comprehensive summary of the recent advances in the study of lncRNAs associated with DLBCL and discuss the clinical relevance of these lncRNAs.
Overview of lncRNAs
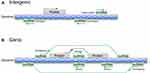
lncRNAs comprise a large and heterogeneous class of transcripts that are greater than 200 nucleotides in length and lack protein-coding capacity. These ncRNAs are generally transcribed and capped by RNA polymerase II, contain introns, are often polyadenylated, and can undergo common posttranscriptional modifications.10 lncRNAs are frequently evolutionarily conserved, intricately regulated and functionally important.11,12 So far, tens of thousands of lncRNAs have been annotated in different databases, such as the NONCODE database (http://www.noncode.org) and LncRBase (http://bicresources.jcbose.ac.in/zhumur/lncrbase/), and the number is continuously increasing.13 The large number and various functions of lncRNAs pose a major challenge to their effective classification. To date, several classification methods have been developed. According to the GENCODE v7 catalog of human lncRNAs, lncRNAs are classified based on their relative positions with respect to protein-coding genes as either 1) intergenic lncRNAs, which are located between two independent genes and can be transcribed from the sense or antisense strand in a divergent or convergent manner, or 2) genic lncRNAs, which can be divided into the following three subtypes: a) exonic lncRNAs, which intersect a protein-coding gene by at least 1 bp; b) intronic lncRNAs, which reside within the intron of a protein-coding gene in either a sense or an antisense orientation; and c) overlapping lncRNAs, which contain a protein-coding gene within an intron in either a sense or an antisense orientation (Figure 1).14,15 Furthermore, recent studies have recognized that lncRNAs are tightly regulated and are expressed in highly lineage-, spatiotemporal-, tissue- and cell type-specific manners.13,16,17
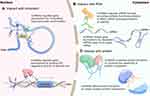
The molecular functions of lncRNAs generally rely on interactions with cellular macromolecules: 1) interactions with chromatin can regulate gene expression by modulating the local chromatin conformation or guiding regulatory molecules to specific loci; 2) interactions with numerous proteins can promote the assembly of protein complexes or impair protein-protein interactions; 3) interactions with mRNAs can affect mRNA splicing, translation or stability through the recruitment of multiple functional proteins; and 4) interactions with miRNAs can shape gene expression by sequestering miRNAs away from their target mRNAs, in agreement with the competing endogenous RNA (ceRNA) hypothesis (Figure 2).18,19 In other words, lncRNAs can function as 1) decoys by “sponging” intracellular molecules to indirectly block their biological functions with downstream factors, 2) guides by recruiting proteins to access regulatory sites of the relevant gene, 3) signals by acting as molecular signals to modulate transcription in response to various stimuli, 4) scaffolds by forming central platforms for the assembly of other molecules or 5) enhancers by binding with mediator complexes to enhance transcription.10,20-22 However, the functions of the vast majority of lncRNAs remain unidentified.
lncRNAs as Candidate Prognostic Biomarkers in DLBCL
Since the development of genomics, numerous methods, including high-throughput sequencing, microarray analysis, and RT-qPCR, have been developed for the detection of lncRNAs. Numerous studies have suggested that aberrant lncRNA expression is closely related to DLBCL. The International Prognostic Index (IPI), which is the most important predictive model for DLBCL, is considered the strongest prognostic indicator and provides the most robust guidance for individual treatment.23 Although the IPI score is currently the most widely used predictive model for DLBCL, it has some limitations.24,25 Numerous studies have attempted to correlate distinct lncRNA expression patterns with clinical diagnoses, subtype classifications, treatment responses and survival outcomes with the aim of highlighting putative prognostic biomarkers for this disease.
Verma et al26 performed a systematic analysis to identify novel lncRNAs from the polyadenylated transcriptome of 116 primary DLBCL samples. These researchers identified 2632 novel lncRNAs expressed in more than one tumor: two-thirds of these were not expressed in normal B cells, and more than one-third of these lncRNAs exhibited differential expression between the two major DLBCL subtypes—activated B-cell (ABC) and germinal center B-cell (GCB). Their study significantly expanded the DLBCL transcriptome and provides an abundant information source for further research. Gao et al27 applied microarray technology to profile the expression of lncRNAs in two different GCB-DLBCL cell lines (OCI-ly1 and OCI-ly19) and normal B lymphocytes, and their data revealed that 1648 lncRNAs were upregulated and 2671 lncRNAs were downregulated in GCB-DLBCL cell lines. In addition, this research group detected the expression levels of five lncRNAs that exhibit significant changes in expression (upregulated: NAALADL2-AS2, HOTAIRM1 and AFAP1-AS1; downregulated: RP4-545C24.1 and OR2A1-AS1) in clinical samples by RT-qPCR and found that the expression trends were similar to those indicated by the microarray data. Another study showed that 1053 lncRNAs were significantly differentially expressed in DLBCL cell lines compared with normal B cells, and 416 and 637 of these lncRNAs were upregulated and downregulated, respectively. Among the differentially expressed lncRNAs, NAALADL2-AS2 exhibited the greatest upregulation, and NONHSAT120161 showed the strongest downregulation.28 Zhou et al29 performed a genome-wide comparative analysis of lncRNA expression profiles in a large number of DLBCL patients from the Gene Expression Omnibus (GEO) database, including patients in three cohorts (total number=905). Their study identified 17 of the 156 differentially expressed lncRNAs between the GCB and ABC subtypes as candidate biomarkers and established a 17-lncRNA signature (ENTPD1-AS1, SACS-AS1, SH3BP5-AS1, RP11-101C11.1, AC009892.10, RP1-68D18.4, MIR600HG, RP11-278 J6.4, RP11-203B7.2, CSMD2-AS1, CTC-467 M3.1, RP4-788P17.1, RP11-553 L6.5, CRNDE, RP11-519G16.3, RP11-21 L19.1 and MME-AS1) for subtype classification (GCB and ABC) and prognosis prediction (overall survival (OS) and progression-free survival (PFS)) that showed favorable reproducible predictive power. Sun et al30 analyzed the lncRNA expression profiles in three cohorts of 1043 DLBCL patients from the GEO database and identified a set of six lncRNAs that were significantly associated with patient OS. The six-lncRNA signature (SACS-AS1, MME-AS1, CSMD2-AS1, RP11-360F5.1, RP1125K19.1, and CTC-467M3.1) can be used as a predictive factor for dividing DLBCL patients with similar IPI scores into a high-risk group and a low-risk group that exhibit significantly different survival outcomes. Another bioinformatic analysis based on available RNA sequencing metadata in cancer bioportals revealed that the lncRNAs GAS5, MIR17HG, HULC and PCA3 were highly altered in DLBCL patients, and among these lncRNAs, abnormal expression of GAS5 was most commonly detected.31 In addition, a meta-analysis conducted by Xu et al demonstrated that lncRNA expression profiling exhibits advantages in the diagnosis and classification of DLBCL. Aberrant expression levels of lncRNAs are also strongly related to worse OS and appear to be promising predictors of DLBCL prognosis.32
Several studies have also demonstrated the prognostic significance of single lncRNAs in DLBCL. The lncRNA HOX transcript antisense RNA (HOTAIR) has been widely confirmed as a crucial regulator in human cancers.33 Recent studies verified that HOTAIR is overexpressed in DLBCL tumor tissues and cell lines and found that this lncRNA is significantly correlated with the tumor sizes, clinical stages, B symptoms and IPI scores of the patients. A higher expression level of HOTAIR is often correlated with a shorter OS time and poorer prognosis and might serve as an independent predictive factor for DLBCL patients.34–36 Shi et al indicated that the lncRNA functional intergenic repeating RNA element (FIRRE) is significantly upregulated in both DLBCL patient samples and cell lines. High FIRRE levels are closely associated with poor OS.37 Another lncRNA, nuclear paraspeckle assembly transcript 1_1 (NEAT1_1), which usually functions as an oncogene in human malignancies, is highly expressed in DLBCL patients and might serve as a predictive marker of poor prognosis.38 Tang et al found that increased levels of the lncRNA RP11-513G11.1 in the peripheral blood of patients with DLBCL predict significantly shortened PFS and OS times.39 By repurposing 1403 normal and DLBCL samples from seven microarray datasets, Zhao et al found that increased expression of the lncRNA NONHSAG026900 might be an independent biomarker of favorable prognosis in DLBCL and can improve the predictive power of the IPI scoring system.40 The results from a study conducted by Wang et al imply that a decreased level of the lncRNA p21-associated ncRNA DNA damage activated (PANDA) in serum is strongly correlated with poorer clinical outcomes and lower OS rates in patients with DLBCL.41 Peng et al found that the lncRNA long intergenic noncoding RNA-p21 (lincRNA-p21) is markedly downregulated in DLBCL and associated with clinical characteristics. Patients with high lincRNA-p21 expression levels exhibit favorable OS and PFS outcomes.42 In addition, several studies have indicated aberrant expression patterns of the lncRNAs small nucleolar RNA host gene 14 (SNHG14), small nucleolar RNA host gene 16 (SNHG16), taurine-upregulated gene 1 (TUG1), metastasis-associated lung adenocarcinoma transcript 1 (MALAT1) and mothers against decapentaplegic homolog 5-antisense RNA 1 (SMAD5-AS1) in DLBCL tumor tissues and cell lines; these findings indicate that these lncRNAs can potentially serve as biomarkers for DLBCL, but this possibility needs further study.43–47
Functional Studies of lncRNAs in DLBCL
Functional studies of numerous lncRNAs that are aberrantly expressed in DLBCL have attempted to shed light on their precise involvement in various aspects of tumor cell biology. These lncRNAs are reportedly extensively involved in most biological mechanisms and pathophysiological processes of DLBCL. For instance, the lncRNA MALAT1 is upregulated in DLBCL and functions as a ceRNA sponge of miR-195 to activate the expression of the immune checkpoint molecule PD-L1, which results in enhanced cell proliferation, migration and immune escape abilities. In addition, MALAT1 can promote CD8+ T cell apoptosis and EMT-like processes (via Ras/ERK signaling).46 Interestingly, MALAT1 overexpression is correlated with chemotherapeutic resistance to adriamycin by enhancing protective cell autophagy.48 The lncRNA SNHG14 elicits oncogenic functions in multiple types of cancers, including DLBCL.49 A recent study showed that increased levels of SNHG14 promote the proliferation, migration and epithelial-to-mesenchymal transition (EMT) of DLBCL cells. Additionally, SNHG14 can act as a ceRNA sponge of miR-5590-3p and subsequently reactivates the downstream protein ZEB1. ZEB1 can transcriptionally activate the inhibitory immunoreceptor PD-L1 to promote tumor cell immune evasion, which suggests that SNHG14 targeting might be a potential strategy for improving the efficacy of DLBCL immunotherapy.43 As another member of the same gene family, SNHG16 can also promote proliferation and cell cycle progression by directly interacting with miR-497-5p and inversely increasing the abundance of the downstream oncoprotein PIM1 in DLBCL cells. Thus, SNHG16 might be a promising therapeutic target in DLBCL.44 According to a study conducted by Zhao et al, the lncRNA SMAD5-AS1 is often downregulated and might act as a tumor suppressor in DLBCL. Further studies have shown that SMAD5-AS1 can inhibit DLBCL cell proliferation by sponging miR-135b-5p to upregulate the expression of downstream APC (which negatively regulates the Wnt/β-catenin pathway by promoting β-catenin degradation50) and inactivate the classical Wnt/β-catenin pathway.47 HOTAIR is one of most studied lncRNAs, and its expression is correlated with poor prognosis in various types of tumors.33 HOTAIR is involved in epigenetic regulation through chromatin remodeling by recruiting polycomb repressive complex 2 (PRC2) proteins (EZH2, SUZ12, and EED) that induce histone H3 trimethylation at lysine 27 (H3K27me3).33 Elevated HOTAIR expression might be strongly related to aggressive DLBCL, possibly by inducing H3K27me3 via EZH2-related PRC2 activation.35 Moreover, Yan et al revealed that the knockdown of HOTAIR induces growth inhibition, cell cycle arrest and apoptosis induction in DLBCL cells, possibly through the PI3K/AKT/NF‑κB pathway.34 The lncRNA highly upregulated in liver cancer (HULC), which was originally found to be strongly overexpressed in human hepatocellular carcinoma,51 is also involved in the pathogenesis of DLBCL.52 Mechanistic data suggest that HULC can maintain cell survival and that the targeting of HULC by β-elemene (a compound derived from the herb Curcuma wenyujin) can effectively suppress cell growth and induce apoptosis in DLBCL.52 Cheng et al demonstrated that the overexpression of the lncRNA TUG1 has oncogenic effects in DLBCL, and this effects are mediated via direct interaction with the oncoprotein MET accompanied by inhibition of its subsequent ubiquitination and degradation. The targeting of TUG1 by gene knockdown can significantly suppress cell proliferation and tumor growth in DLBCL.45 Shi et al focused on the lncRNA FIRRE, which can activate the Wnt/β-catenin signaling pathway to facilitate DLBCL cell growth through the modulation of β-catenin nuclear translocation. This research group also demonstrated that the MYC proto-oncogene, which is often highly expressed in DLBCL, contributes to the transcriptional activation of FIRRE.37 The MAPK/ERK pathway plays an important role in modulating cell proliferation and the cell cycle in tumors, and functional interactions exist between p53 and MAPK/ERK signaling.53 A study conducted by Wang et al revealed that the lncRNA PANDA is activated by p53 and functions as a tumor suppressor gene through the inhibition of cell growth via G0/G1 cell cycle arrest by inactivating the MAPK/ERK pathway in DLBCL cells. In addition, treatment with the MAPK/ERK agonist anisomycin can potently abolish the antitumor effect of PANDA.41 Previous studies have shown that the lncRNA NEAT1_1 is highly expressed and predicts poor prognosis in DLBCL, and the knockdown of NEAT1_1 results in growth inhibition, cell cycle arrest and apoptosis induction in tumor cells.38 In contrast, the lncRNA lincRNA-p21 (which is downregulated in DLBCL and acts as a tumor suppressor) can inhibit cell proliferation and induce cell cycle arrest by functionally modulating downstream p21, cyclin D1 and CDK4 expression in DLBCL cells.42 Moreover, through a bioinformatic analysis, Tian et al revealed that some differentially expressed lncRNAs (protein kinase C theta-antisense RNA 1 (PRKCQ-AS1), human leukocyte antigen complex P5 (HCP5), OPA-interacting protein 5-antisense RNA 1 (OIP5-AS1), growth arrest-specific 5 (GAS5) and TUG1) might perform crucial functions in the transformation of DLBCL by regulating a series of biological processes, including antiapoptotic processes, the cell cycle, DNA repair, oxidative stress responses and transcription.54
DLBCL-related lncRNAs and their roles in DLBCL are summarized in Table 1.
 |
Table 1 Examples of lncRNAs with Prognostic and/or Functional Impact in DLBCL |
Conclusions and Perspectives
The study of lncRNAs in human malignancies, including DLBCL, remains in its infancy. However, high-throughput sequencing technologies and bioinformatic analysis approaches are increasing the possibility of identifying lncRNAs that are potentially involved in DLBCL. An increasing body of data indicates that lncRNAs play a critical role in the development and pathogenesis of DLBCL, and the lncRNA expression profiles of DLBCL patients might be used for their diagnosis and classification, the prediction of their prognoses and the establishment of targeted therapies. In this review, we discuss the lncRNAs associated with DLBCL, summarize their characteristics and molecular functions, and describe their potential roles as biomarkers.
Based on published studies, lncRNAs are crucial players in regulating multiple oncogenic genes or signaling pathways across DLBCL through multiple aspects of epigenetic regulation. The specific expression patterns of lncRNAs in DLBCL also make them good candidates as diagnostic biomarkers or therapeutic targets. For instance, the well-studied lncRNAs HOTAIR and MALAT-1 have been shown to play important prognostic roles and might serve as potential targets for therapeutic intervention in DLBCL.34–36,46,48 However, the relevant studies related to DLBCL remain limited, and the available data are not sufficient for conclusively demonstrating that a single lncRNA is representative, highly sensitive, and highly specific and can be used in clinical practice as a biomarker. The combination of several lncRNAs might be more effective than a single one, and this finding, although hypothetical, is supported by the studies conducted by Zhou et al and Sun et al, who established 17-lncRNA and six-lncRNA panels.29,30 Based on the existing studies, we believe that the combination of specific expression patterns of lncRNAs with clinical and genetic prognosticators of DLBCL patients will further improve the accuracy and specificity of individual prognostic assessments. Therefore, more research is needed to obtain further useful information.
Researchers are attempting to develop therapeutic strategies involving the targeting of lncRNAs. As mentioned in our manuscript, many studies have shown favorable in vitro data for targeting aberrantly expressed lncRNAs in DLBCL cells. Additionally, the sensitivity to the conventional standard therapy consisting of R-CHOP might be increased by artificial modulation of lncRNAs expressions. Studies have also attempted to develop small-molecule inhibitors that specifically disrupt lncRNA-protein interactions.55 However, these therapeutic agents that can be safely used for the treatment of DLBCL patients are only beginning to be established. Further studies aiming to identify the most promising lncRNA targets and design the best therapeutics with appropriate efficacy and safety are urgently needed. In summary, we believe that aberrant regulation of lncRNAs in DLBCL is not simply an epiphenomenon but also has great potential for the identification of biomarkers and therapeutic targets. We anticipate that further in-depth investigation of the roles of lncRNAs in DLBCL might help unravel their intricate regulatory networks and provide important insights into DLBCL treatment.
Abbreviations
DLBCL, diffuse large B-cell lymphoma; lncRNAs, long noncoding RNAs; NHL, non-Hodgkin lymphoma; ncRNAs, noncoding RNAs; miRNAs, microRNAs; circRNAs, circular RNAs; ceRNA, competing endogenous RNA; IPI, International Prognostic Index; GEO, Gene Expression Omnibus; OS, overall survival; PFS, progression-free survival; EMT, epithelial-to-mesenchymal transition; PRC2, polycomb repressive complex 2; H3K27me3, H3 trimethylation at lysine 27; HOTAIR, HOX transcript antisense RNA; FIRRE, functional intergenic repeating RNA element; NEAT1_1, nuclear paraspeckle assembly transcript 1_1; PANDA, p21 associated ncRNA DNA damage activated; HULC, highly upregulated in liver cancer; lincRNA-p21, long intergenic noncoding RNA-p21; SNHG14, small nucleolar RNA host gene 14; SNHG16, small nucleolar RNA host gene 16; TUG1, taurine-upregulated gene 1; MALAT1, metastasis-associated lung adenocarcinoma transcript 1; SMAD5-AS1, mothers against decapentaplegic homolog 5-antisense RNA 1; PRKCQ-AS1, protein kinase C theta-antisense RNA 1; HCP5, human leukocyte antigen complex P5; OIP5-AS1, OPA-interacting protein 5-antisense RNA 1; GAS5, growth arrest-specific 5.
Acknowledgments
We are grateful for the financial support provided by the Natural Science Foundation of China (No. 81900152), the Natural Science Foundation of Zhejiang Province (No. LQ19H080005), and the Health Department of Zhejiang Province (No. 2020KY113).
Disclosure
The authors declare that they have no competing interests.
References
1. Pasqualucci L, Dalla-Favera R. Genetics of diffuse large B-cell lymphoma. Blood. 2018;131(21):2307–2319. doi:10.1182/blood-2017-11-764332
2. Smith A, Howell D, Patmore R, Jack A, Roman E. Incidence of haematological malignancy by sub-type: a report from the Haematological Malignancy Research Network. Br J Cancer. 2011;105(11):1684–1692. doi:10.1038/bjc.2011.450
3. Feugier P, Van Hoof A, Sebban C, et al. Long-term results of the R-CHOP study in the treatment of elderly patients with diffuse large B-cell lymphoma: a study by the Groupe d’Etude des Lymphomes de l’Adulte. J Clin Oncol. 2005;23(18):4117–4126. doi:10.1200/JCO.2005.09.131
4. Roschewski M, Staudt LM, Wilson WH. Diffuse large B-cell lymphoma-treatment approaches in the molecular era. Nat Rev Clin Oncol. 2014;11(1):12–23. doi:10.1038/nrclinonc.2013.197
5. Coiffier B, Sarkozy C. Diffuse large B-cell lymphoma: R-CHOP failure-what to do? Hematology Am Soc Hematol Educ Program. 2016;2016(1):366–378. doi:10.1182/asheducation-2016.1.366
6. Wright MW, Bruford EA. Naming ‘junk’: human non-protein coding RNA (ncRNA) gene nomenclature. Hum Genomics. 2011;5(2):90–98. doi:10.1186/1479-7364-5-2-90
7. Anastasiadou E, Jacob LS, Slack FJ. Non-coding RNA networks in cancer. Nat Rev Cancer. 2018;18(1):5–18. doi:10.1038/nrc.2017.99
8. Spizzo R, Almeida MI, Colombatti A, Calin GA. Long non-coding RNAs and cancer: a new frontier of translational research? Oncogene. 2012;31(43):4577–4587. doi:10.1038/onc.2011.621
9. Bhan A, Soleimani M, Mandal SS. Long noncoding RNA and cancer: a new paradigm. Cancer Res. 2017;77(15):3965–3981. doi:10.1158/0008-5472.CAN-16-2634
10. Quinn JJ, Chang HY. Unique features of long non-coding RNA biogenesis and function. Nat Rev Genet. 2016;17(1):47–62. doi:10.1038/nrg.2015.10
11. Guttman M, Amit I, Garber M, et al. Chromatin signature reveals over a thousand highly conserved large non-coding RNAs in mammals. Nature. 2009;458(7235):223–227. doi:10.1038/nature07672
12. Hezroni H, Koppstein D, Schwartz MG, Avrutin A, Bartel DP, Ulitsky I. Principles of long noncoding RNA evolution derived from direct comparison of transcriptomes in 17 species. Cell Rep. 2015;11(7):1110–1122. doi:10.1016/j.celrep.2015.04.023
13. Iyer MK, Niknafs YS, Malik R, et al. The landscape of long noncoding RNAs in the human transcriptome. Nat Genet. 2015;47(3):199–208. doi:10.1038/ng.3192
14. Harrow J, Frankish A, Gonzalez JM, et al. GENCODE: the reference human genome annotation for The ENCODE Project. Genome Res. 2012;22(9):1760–1774. doi:10.1101/gr.135350.111
15. Derrien T, Johnson R, Bussotti G, et al. The GENCODE v7 catalog of human long noncoding RNAs: analysis of their gene structure, evolution, and expression. Genome Res. 2012;22(9):1775–1789. doi:10.1101/gr.132159.111
16. Cabili MN, Trapnell C, Goff L, et al. Integrative annotation of human large intergenic noncoding RNAs reveals global properties and specific subclasses. Genes Dev. 2011;25(18):1915–1927. doi:10.1101/gad.17446611
17. Li F, Xiao Y, Huang F, et al. Spatiotemporal-specific lncRNAs in the brain, colon, liver and lung of macaque during development. Mol Biosyst. 2015;11(12):3253–3263. doi:10.1039/C5MB00474H
18. Guttman M, Rinn JL. Modular regulatory principles of large non-coding RNAs. Nature. 2012;482(7385):339–346. doi:10.1038/nature10887
19. Salmena L, Poliseno L, Tay Y, Kats L, Pandolfi PP. A ceRNA hypothesis: the rosetta stone of a hidden RNA language? Cell. 2011;146(3):353–358. doi:10.1016/j.cell.2011.07.014
20. Jeon Y, Lee JT. YY1 tethers Xist RNA to the inactive X nucleation center. Cell. 2011;146(1):119–133. doi:10.1016/j.cell.2011.06.026
21. Rinn JL, Chang HY. Genome regulation by long noncoding RNAs. Annu Rev Biochem. 2012;81(1):145–166. doi:10.1146/annurev-biochem-051410-092902
22. Ørom UA, Derrien T, Beringer M, et al. Long noncoding RNAs with enhancer-like function in human cells. Cell. 2010;143(1):46–58. doi:10.1016/j.cell.2010.09.001
23. International Non-Hodgkin’s Lymphoma Prognostic Factors Project. A predictive model for aggressive non-hodgkin’s lymphoma. N Engl J Med. 1993;329(14):987–994. doi:10.1056/NEJM199309303291402
24. Clausen MR, Maurer MJ, Ulrichsen SP, et al. Pretreatment hemoglobin adds prognostic information to the NCCN-IPI in patients with diffuse large B-cell lymphoma treated with anthracycline-containing chemotherapy. Clin Epidemiol. 2019;11:987–996. doi:10.2147/CLEP.S219595
25. Goto N, Tsurumi H, Kasahara S, et al. Serum interleukin-18 level is associated with the outcome of patients with diffuse large B-cell lymphoma treated with CHOP or R-CHOP regimens. Eur J Haematol. 2011;87(3):217–227. doi:10.1111/j.1600-0609.2011.01649.x
26. Verma A, Jiang Y, Du W, Fairchild L, Melnick A, Elemento O. Transcriptome sequencing reveals thousands of novel long non-coding RNAs in B cell lymphoma. Genome Med. 2015;7(1):110. doi:10.1186/s13073-015-0230-7
27. Gao HY, Wu B, Yan W, et al. Microarray expression profiles of long non-coding RNAs in germinal center-like diffuse large B-cell lymphoma. Oncol Rep. 2017;38(3):1363–1372. doi:10.3892/or.2017.5821
28. Zhu D, Fang C, Li X, et al. Predictive analysis of long non-coding RNA expression profiles in diffuse large B-cell lymphoma. Oncotarget. 2017;8(14):23228–23236. doi:10.18632/oncotarget.15571
29. Zhou M, Zhao H, Xu W, Bao S, Cheng L, Sun J. Discovery and validation of immune-associated long non-coding RNA biomarkers associated with clinically molecular subtype and prognosis in diffuse large B cell lymphoma. Mol Cancer. 2017;16(1):16. doi:10.1186/s12943-017-0580-4
30. Sun J, Cheng L, Shi H, et al. A potential panel of six-long non-coding RNA signature to improve survival prediction of diffuse large-B-cell lymphoma. Sci Rep. 2016;6(1):27842. doi:10.1038/srep27842
31. Dousti F, Shahrisa A, Ansari H, et al. Long non-coding RNAs expression levels in diffuse large B-cell lymphoma: an in silico analysis. Pathol Res Pract. 2018;214(9):1462–1466. doi:10.1016/j.prp.2018.08.006
32. Xu P, Chen X, Su P. A pooled analysis of the clinical utilities of long non-coding RNA based molecular signature for diffuse large B cell lymphoma. Clin Lab. 2017;63(11+12/2017):1831–1840. doi:10.7754/Clin.Lab.2017.170605
33. Gupta RA, Shah N, Wang KC, et al. Long non-coding RNA HOTAIR reprograms chromatin state to promote cancer metastasis. Nature. 2010;464(7291):1071–1076. doi:10.1038/nature08975
34. Yan Y, Han J, Li Z, Yang H, Sui Y, Wang M. Elevated RNA expression of long non‑coding HOTAIR promotes cell proliferation and predicts a poor prognosis in patients with diffuse large B cell lymphoma. Mol Med Rep. 2016;13(6):5125–5131. doi:10.3892/mmr.2016.5190
35. Oh EJ, Kim SH, Yang WI, Ko YH, Yoon SO. Long non-coding RNA HOTAIR expression in diffuse large B-cell lymphoma: in relation to polycomb repressive complex pathway proteins and H3K27 trimethylation. J Pathol Transl Med. 2016;50(5):369–376. doi:10.4132/jptm.2016.06.06
36. Lin Y, Fang Z, Lin Z, et al. The prognostic impact of long noncoding RNA HOTAIR in leukemia and lymphoma: a meta-analysis. Hematology. 2018;23(9):600–607. doi:10.1080/10245332.2018.1446572
37. Shi X, Cui Z, Liu X, et al. LncRNA FIRRE is activated by MYC and promotes the development of diffuse large B-cell lymphoma via Wnt/β-catenin signaling pathway. Biochem Biophys Res Commun. 2019;510(4):594–600. doi:10.1016/j.bbrc.2019.01.105
38. Deng L, Jiang L, Tseng KF, et al. Aberrant NEAT1_1 expression may be a predictive marker of poor prognosis in diffuse large B cell lymphoma. Cancer Biomark. 2018;23(2):157–164. doi:10.3233/CBM-160221
39. Tang JL, Li XM, Zhang L. Expression and significance of lncRNA RP11-513G11.1 in peripheral blood of patients with diffuse large B-cell lymphoma. Zhongguo Shi Yan Xue Ye Xue Za Zhi. 2019;27(5):1515–1521. doi:10.19746/j.cnki.issn.1009-2137.2019.05.024
40. Zhao S, Fang S, Liu Y, et al. The long non-coding RNA NONHSAG026900 predicts prognosis as a favorable biomarker in patients with diffuse large B-cell lymphoma. Oncotarget. 2017;8(21):34374–34386. doi:10.18632/oncotarget.16163
41. Wang Y, Zhang M, Xu H, et al. Discovery and validation of the tumor-suppressive function of long noncoding RNA PANDA in human diffuse large B-cell lymphoma through the inactivation of MAPK/ERK signaling pathway. Oncotarget. 2017;8(42):72182–72196. doi:10.18632/oncotarget.20053
42. Peng W, Wu J, Feng J. LincRNA-p21 predicts favorable clinical outcome and impairs tumorigenesis in diffuse large B cell lymphoma patients treated with R-CHOP chemotherapy. Clin Exp Med. 2017;17(1):1–8. doi:10.1007/s10238-015-0396-8
43. Zhao L, Liu Y, Zhang J, Liu Y, Qi Q. LncRNA SNHG14/miR-5590-3p/ZEB1 positive feedback loop promoted diffuse large B cell lymphoma progression and immune evasion through regulating PD-1/PD-L1 checkpoint. Cell Death Dis. 2019;10(10):731. doi:10.1038/s41419-019-1886-5
44. Zhu Q, Li Y, Guo Y, et al. Long non-coding RNA SNHG16 promotes proliferation and inhibits apoptosis of diffuse large B-cell lymphoma cells by targeting miR-497-5p/PIM1 axis. J Cell Mol Med. 2019;23(11):7395–7405. doi:10.1111/jcmm.14601
45. Cheng H, Yan Z, Wang X, et al. Downregulation of long non-coding RNA TUG1 suppresses tumor growth by promoting ubiquitination of MET in diffuse large B-cell lymphoma. Mol Cell Biochem. 2019;461(1–2):47–56. doi:10.1007/s11010-019-03588-7
46. Wang QM, Lian GY, Song Y, Huang YF, Gong Y. LncRNA MALAT1 promotes tumorigenesis and immune escape of diffuse large B cell lymphoma by sponging miR-195. Life Sci. 2019;231:116335. doi:10.1016/j.lfs.2019.03.040
47. Zhao CC, Jiao Y, Zhang YY, et al. Lnc SMAD5-AS1 as ceRNA inhibit proliferation of diffuse large B cell lymphoma via Wnt/β-catenin pathway by sponging miR-135b-5p to elevate expression of APC. Cell Death Dis. 2019;10(4):252. doi:10.1038/s41419-019-1479-3
48. Li LJ, Chai Y, Guo XJ, Chu SL, Zhang LS. The effects of the long non-coding RNA MALAT-1 regulated autophagy-related signaling pathway on chemotherapy resistance in diffuse large B-cell lymphoma. Biomed Pharmacother. 2017;89:939–948. doi:10.1016/j.biopha.2017.02.011
49. Han Y, Zhou S, Wang X, Mao E, Huang L. SNHG14 stimulates cell autophagy to facilitate cisplatin resistance of colorectal cancer by regulating miR-186/ATG14 axis. Biomed Pharmacother. 2020;121:109580. doi:10.1016/j.biopha.2019.109580
50. Qian J, Huang X, Zhang Y, Ye X, Qian W. γ-catenin overexpression in AML patients may promote tumor cell survival via activation of the Wnt/β-catenin axis. Onco Targets Ther. 2020;13:1265–1276. doi:10.2147/OTT.S230873
51. Zhang H, Liao Z, Liu F, et al. Long noncoding RNA HULC promotes hepatocellular carcinoma progression. Aging (Albany NY). 2019;11(20):9111–9127. doi:10.18632/aging.102378
52. Hu T, Gao Y. β-elemene suppresses tumor growth of diffuse large B-cell lymphoma through regulating lncRNA HULC-mediated apoptotic pathway. Biosci Rep. 2020;40(2):BSR20190804. doi:10.1042/BSR20190804
53. Wu GS. The functional interactions between the p53 and MAPK signaling pathways. Cancer Biol Ther. 2004;3(2):156–161. doi:10.4161/cbt.3.2.614
54. Tian L, He Y, Zhang H, Wu Z, Li D, Zheng C. Comprehensive analysis of differentially expressed profiles of lncRNAs and mRNAs reveals ceRNA networks in the transformation of diffuse large B-cell lymphoma. Oncol Lett. 2018;16(1):882–890. doi:10.3892/ol.2018.8722
55. Pedram Fatemi R, Salah-Uddin S, Modarresi F, Khoury N, Wahlestedt C, Faghihi MA. Screening for small-molecule modulators of long noncoding RNA-protein interactions using AlphaScreen. J Biomol Screen. 2015;20(9):1132–1141. doi:10.1177/1087057115594187
 © 2020 The Author(s). This work is published and licensed by Dove Medical Press Limited. The full terms of this license are available at https://www.dovepress.com/terms.php and incorporate the Creative Commons Attribution - Non Commercial (unported, v3.0) License.
By accessing the work you hereby accept the Terms. Non-commercial uses of the work are permitted without any further permission from Dove Medical Press Limited, provided the work is properly attributed. For permission for commercial use of this work, please see paragraphs 4.2 and 5 of our Terms.
© 2020 The Author(s). This work is published and licensed by Dove Medical Press Limited. The full terms of this license are available at https://www.dovepress.com/terms.php and incorporate the Creative Commons Attribution - Non Commercial (unported, v3.0) License.
By accessing the work you hereby accept the Terms. Non-commercial uses of the work are permitted without any further permission from Dove Medical Press Limited, provided the work is properly attributed. For permission for commercial use of this work, please see paragraphs 4.2 and 5 of our Terms.