Back to Journals » OncoTargets and Therapy » Volume 9
Long noncoding RNA ZNFX1-AS1 suppresses growth of hepatocellular carcinoma cells by regulating the methylation of miR-9
Authors Wang T, Ma S, Qi X, Tang X, Cui D, Wang Z, Chi J, Li P, Zhai B
Received 29 December 2015
Accepted for publication 29 March 2016
Published 12 August 2016 Volume 2016:9 Pages 5005—5014
DOI https://doi.org/10.2147/OTT.S103329
Checked for plagiarism Yes
Review by Single anonymous peer review
Peer reviewer comments 2
Editor who approved publication: Dr William C. Cho
Tao Wang, Sicong Ma, Xingxing Qi, Xiaoyin Tang, Dan Cui, Zhi Wang, Jiachang Chi, Ping Li, Bo Zhai
Department of Interventional Oncology, Renji Hospital, School of Medicine, Shanghai Jiao Tong University, Shanghai, People’s Republic of China
Abstract: Many long noncoding RNAs have been reported to play pivotal roles in cancer biology. Among them, the long noncoding RNA ZNFX1-AS1 has been confirmed to function in breast cancer progression, but the role of ZNFX1-AS1 in hepatocellular carcinoma (HCC) growth and the related molecular mechanisms still remains unknown. In the present study, we first identified the expression of ZNFX1-AS1 in HCC patients’ specimens and HCC cell lines through quantitative reverse transcription polymerase chain reaction. Next, the effects of ZNFX1-AS1 on HCC cell growth and apoptosis were analyzed. MTT assay was used to measure the cell numbers, and fluorescence-activated cell sorting analysis was performed to evaluate cell apoptosis. Finally, the relationship between ZNFX1-AS1 and miR-9 in HCC was studied. Our results suggest that ZNFX1-AS1 was markedly downregulated in HCC samples and cell lines. Overexpression of ZNFX1-AS1 inhibited the cell proliferation and colony formation in HCC cell lines and also induced HCC cell apoptosis. Additionally, miR-9 was lowly expressed in HCC tissues and positively correlated with ZNFX1-AS1 expression. Meanwhile, significant upregulation of miR-9 and downregulation of the methylation of miR-9 promoter CpG island were observed when ZNFX1-AS1 was overexpressed. In summary, our results indicate that ZNFX1-AS1 plays a vital role in HCC progression via regulating the methylation of miR-9 and may be a potential tumor suppressor.
Keywords: ZNFX1-AS1, hepatocellular carcinoma, miR-9, methylation
Introduction
Hepatocellular carcinoma (HCC) has become the most common leading cause of cancer-related mortality globally.1 Despite increasing exploration for novel treatments, such as operation, liver transplantation, and chemoradiotherapy, there has still been little success in improving the 5-year overall survival (OS) rate of HCC patients.2,3 The poor understanding of the molecular mechanisms of development and progression of HCC is one of the most important reasons that result in high fatality and poor prognosis.4,5 Therefore, probing the molecular mechanisms underlying occurrence of HCC is of great clinical value for developing new preventable and therapeutic strategies.
Large-scale researches on long noncoding RNAs (lncRNAs), non-protein-coding transcripts that are more than 200 nt in length, have shown that lncRNAs are involved in a variety of biological processes, including silencing the X chromosome, transcriptional regulation, modifying chromatin, splicing, and so on.6–12 Recently, numerous specifically dysregulated lncRNAs have been found in several cancer types,13 which can be seen as a potential clinical advantage as these lncRNAs can function as diagnostic or prognostic biomarkers.14,15 LncRNAs, such as anti-NOS2A16 and MEG3,17–19 have been identified to be associated with the pathogenesis of brain tumor. In addition, H19,20 BC200,21,22 HOTAIR,23 and MALAT124 have been proven to play crucial roles in the pathological process of breast tumor. Since the lncRNA HULC was reported to be specifically upregulated in HCC,25 an increasing number of lncRNAs have been found to function in HCC progression, such as MALAT1, PCNA-AS1, and HOTTIP.26–28 ZNFX1-AS1, an lncRNA that transcripts from the antisense strand near the 5′-end of the protein-coding gene Znfx1, hosts three C/D box snoRNAs (SNORDs): Snord12, Snord12b, and Snord12c.29 ZNFX1-AS1 has been reported to be related to several cancers, including breast, gastric, and colorectal cancer.30 In addition, ZNFX1-AS1 is significantly downregulated in breast cancer, and hence can function as a potential biomarker for breast cancer.29 Nevertheless, its role in other cancers remains largely unknown, especially in HCC.
In this study, we identified the significant downregulation of ZNFX1-AS1 in HCC tissues, which was associated with the proliferation and apoptosis of HCC cells. Moreover, ZNFX1-AS1 was also found to function in the progression of HCC via regulating the methylation of miR-9.
Materials and methods
HCC specimens and cell lines
Eighty-four pairs of samples (including 84 HCC samples and normal adjacent liver tissues) from HCC patients were obtained from Eastern Hepatobiliary Surgery Hospital (Shanghai, People’s Republic of China). The study was approved by the Ethics Committee of the Eastern Hepatobiliary Surgery Hospital. Written informed consent was obtained for the use of tissue samples from all patients. None of the patients recruited for this study had anticancer treatments before the surgery. Liver cancer cell lines HepG2, Hep3B, SNU449, HuH-7, and normal human hepatocytes (HH) were purchased from American Type Culture Collection (ATCC, Manassas, VA, USA). The earlier cell lines were cultured in high-glucose Dulbecco’s Modified Eagle’s Medium (Thermo Fisher Scientific, Waltham, MA, USA), supplemented with 10% fetal bovine serum, 100 U/mL penicillin, and 100 mg/mL streptomycin in a humidified incubator with 5% CO2 at 37°C.
Quantitative reverse transcription polymerase chain reaction and methylation-specific polymerase chain reaction
Total RNA from HCC tissues was extracted using Trizol reagent (Thermo Fisher Scientific). Real-time polymerase chain reaction (PCR) was performed using the SYBR Green Real-Time PCR Master Mix (TaKaRa, Tokyo, Japan) as described.28 To examine the expression of miR-9, we performed quantitative PCR using synthetic micro-RNA sequences in a calibration curve. The following primers were used:
Forward: 5′-GGGTCTTTGGTTATCTAGC-3′; Reverse: 5′-TGCGTGTCGTGGAGTC3-3′.
Methylation-specific polymerase chain reaction (MSP) was performed as previously described.31 The expression of ZNFX1-AS1 was normalized with GAPDH. The primers of ZNFX1-AS1 and GAPDH were bought from Thermo Fisher Scientific. The primers used for miR-9 MSP in this study are listed as follows:
miR-9-msp-U forward: 5′-GTGAGATTTGTTTTATTTTGTTTGT-3′; Reverse: 5′-AAAACCCTAAATTCCAAATACCAA-3′; miR-9-msp-M forward: 5′-AGGTGAGATTCGTTTTATTTTGTTC-3′; Reverse: 5′-AAAACCCTAAATTCCAAATACCG-3′.
Vector construction and transfection
Knockdown lentivirus and the relative negative control lentivirus were purchased from GenePharma (Shanghai, People’s Republic of China). SNU449 and HepG2 cells were transfected with siRNA lentivirus according to the manufacturer’s instructions. Stable transfection lentivirus cell lines were used for subsequent assays. The cDNA encoding ZNFX1-AS1 was PCR-amplified by the PfuUltra II Fusion HS DNA Polymerase (Stratagene, Agilent Technologies, Santa Clara, CA, USA) and subcloned into a pcDNA vector (Invitrogen), named pcDNA-ZNFX1-AS1.
Cell proliferation and apoptosis assay
The cells were seeded in 96-well plates for cell proliferation assay using a 3-(4, 5-dimethylthiazol-2-yl)-2,5-diphenyltetrazolium bromide (MTT) Cell Proliferation/Viability Assay Kit (Sigma-Aldrich Chemie Gmbh, Munich, Germany) in accordance with the guidelines. The cell apoptosis was measured using an Annexin-V-Fluos and Propidium Iodide (PI) Apoptosis Detection Kit (Sigma) by a flow cytometer (Becton Dickinson, San Jose, CA, USA) according to the manufacturer’s instructions.
Soft sugar colony formation assay
The colony number of HepG2 and SNU449 cells was recorded after 2 weeks of soft sugar colony formation analysis. Briefly, cells were suspended in 1.5 mL complete medium supplemented with 0.45% agarose (Thermo Fisher Scientific), and then placed in 35 mm tissue culture plates containing 1.5 mL complete medium and agarose (0.75%) on the bottom layer. Finally, cell colonies were stained with 0.005% crystal violet and analyzed using a microscope.
Western blotting
Western blotting analysis was performed as described in the study by Zhang et al.32 Thirty-microgram total protein was extracted and separated by 10% sodium dodecyl sulfate polyacrylamide gel electrophoresis (SDS-PAGE) gel, and then transferred to polyvinylidene difluoride membranes (Millipore, Darmstadt, Germany). The membrane was blocked with specific primary antibodies and then incubated with secondary antibodies labeled with horseradish peroxidase and detected by chemiluminescence (Thermo, Rockford, IL, USA). DNMT1 and GAPDH antibodies were obtained from Santa Cruz Biotechnology, Santa Cruz, CA, USA and GAPDH was used as protein loading control.
Statistical analysis
Data were presented as mean ± standard deviation (SD) and analyzed using SPSS 17.0 software (SPSS Inc., Chicago, IL, USA). Statistical differences between two independent groups were determined using unpaired t-test. For multiple comparison, one-way analysis of variance was used. Linear regression analysis was performed to analyze the correlation between expressions of miR-9 and ZNFX1-AS1. The Kaplan–Meier method was used to estimate the OS rate. Survival differences according to hMOF expression were analyzed using the log-rank test. P<0.05 was considered statistically significant.
Results
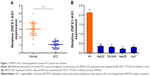
Expression of ZNFX1-AS1 is significantly downregulated in HCC samples and cell lines
We measured ZNFX1-AS1 expression in HCC tissues and cell lines through quantitative PCR. As shown in Figure 1A, compared to normal liver tissues, the expression of ZNFX1-AS1 was prominently decreased in HCC tissues. Remarkably, consistent with the data derived from HCC samples, the expression of ZNFX1-AS1 was significantly decreased in all cell lines (HepG2, Hep3B, SNU449, HuH7) except in normal liver cell line HH (Figure 1B). In summary, ZNFX1-AS1 was downregulated in HCC tissues and cell lines. We selected HepG2 and SNU449 (the relatively higher expression of ZNFX1-AS1) as representatives for further research.
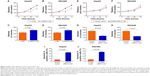
Low expression of ZNFX1-AS1 is associated with poor prognosis of HCC
To analyze the relationship between ZNFX1-AS1 expression and HCC patients’ prognosis, we detected rates of OS and disease-free survival of patients using Kaplan–Maier analysis. As shown in Figure 2A, the 5-year OS rate of patients with high expression of ZNFX1-AS1 was significantly higher than that for the low ZNFX1-AS1 expression. In consistence, the 5-year disease-free survival rate in high ZNFX1-AS1 expression group was also higher than that in the low expression group (Figure 2B).
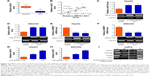
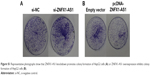
ZNFX1-AS1 suppresses cell proliferation and promotes cell apoptosis in HCC cells
To identify the effects of ZNFX1-AS1 on proliferation and apoptosis of HCC cells, we first determined whether ZNFX1-AS1 siRNA inhibits the expression of ZNFX1-AS1 in HepG2 and SNU449 cells. The results revealed that cell proliferation and colony formation of HepG2 and SNU449 cells were increased when treated with si-ZNFX1-AS1 (Figures 3A–D; S1A). Next, ZNFX1-AS1 was overexpressed to further identify the relationship between ZNFX1-AS1 and cell proliferation and colony formation of HepG2 and SNU449 cells. Our results suggested that ZNFX1-AS1 overexpression inhibited cell proliferation and colony formation of HepG2 and SNU449 cells (Figures 3E–H; S1B). Furthermore, we investigated the effect of ZNFX1-AS1 on cancer cell apoptosis by performing fluorescence-activated cell sorting assay. We found that ZNFX1-AS1 overexpression induced cancer cell apoptosis (Figures 3I and J; S2A and B). Collectively, these results suggest that ZNFX1-AS1 inhibits HCC cell proliferation and promotes cell apoptosis.
ZNFX1-AS1 induces miR-9 expression through inhibiting methylation of CpG island at its promoter
Furthermore, we aimed to explore the mechanisms underlying the potent effect of ZNFX1-AS1 on HCC. Since miR-9 has been reported to be involved in HCC progression,33 we hypothesized that ZNFX1-AS1 may regulate HCC growth by miR-9. Firstly, we examined the expression level of miR-9 in HCC tissues. Compared to the normal tissues, the expression of miR-9 was markedly downregulated in HCC tissues (Figure 4A). Moreover, ZNFX1-AS1 expression level was positively related to miR-9 expression level in HCC tissues (Figure 4B), which implied that ZNFX1-AS1 promoted miR-9 expression in HCC. Next, we explored the relationship between ZNFX1-AS1 and miR-9. As shown in Figure 4C and D, miR-9 was significantly upregulated in HepG2 and SNU449 cells when ZNFX1-AS1 was overexpressed. To further investigate whether ZNFX1-AS1 regulates HCC growth through miR-9, we examined the methylation of miR-9 promoter in HepG2 and SNU449 cells. DNMT1, a kind of methyltransferase, can regulate miRNA expression by controlling methylation of CpG island. Additionally, it was found that ZNFX1-AS1 overexpression inhibited DNMT1 expression (Figures 4E and F; S3). At the same time, the methylation inhibitor Aza increased the expression of miR-9 in HepG2 and SUN449 cells (Figure 4G and H). Indeed, we found that DNMT1 overexpression inhibited the expression of miR-9 in HepG2 cells (Figure S4). Finally, MSP analysis was used to analyze the methylation of miR-9 CpG sites in HepG2, SNU449, Hep3B, and Huh7 cells. Obviously, the overexpression of ZNFX1-AS1 inhibited the methylation of miR-9 CpG island (Figure 4I). Taken together, these results demonstrated that ZNFX1-AS1 may function in HCC progression by regulating the methylation of miR-9 CpG island.
Discussion
A better understanding of the mechanisms underlying HCC metastasis is of importance. Presently, numerous lncRNAs are found to play crucial roles in the metastasis and invasion of HCC cells.34,35 Genetic variants at 20q13 are associated with some tumors.36,37 LncRNA ZNFX1-AS1 is located in the 20q13.13 region, which functions in breast cancer progression, implying a potential role of lncRNA ZNFX1-AS1 in HCC metastasis.30 However, the biological function or molecular mechanisms of ZNFX1-AS1 in HCC remain unclear. Therefore, understanding the effects of ZNFX1-AS1 on HCC is essential.
Here, we identified a novel ZNFX1-AS1-dependent molecular mechanism during the progression of HCC. We reported that ZNFX1-AS1 was prominently downregulated in HCC tissues and liver cancer cell lines. ZNFX1-AS1 low expression was associated with poor prognosis. To explore the functional role of ZNFX1-AS1 in HCC progression, we adopted lentivirus to knockdown or overexpress ZNFX1-AS1 in SNU449 and HepG2 cells. ZNFX1-AS1 was confirmed to function in the proliferation, colony formation, and apoptosis of HCC cells. Our results revealed that ZNFX1-AS1 may play a pivotal role in the growth of HCC.
As miR-9 has been identified to play a role in HCC growth,32 we tried to seek the correlations between ZNFX1-AS1 and miR-9 in HCC progression. In this article, we identified that the expression of miR-9 was downregulated in HCC tissues and positively correlated with the expression of ZNFX1-AS1. More importantly, miR-9 was markedly upregulated in HepG2 and SNU449 cells with ZNFX1-AS1 overexpression, implying that miR-9 may be a target of ZNFX1-AS1 in HCC cells. To further confirm whether ZNFX1-AS1 regulated HCC growth through miR-9, we examined the methylation of miR-9 in HepG2 and SNU449 cells, respectively. Obviously, the overexpression of ZNFX1-AS1 inhibited the methylation of miR-9 CpG island. Conclusively, these results indicated that ZNFX1-AS1 may function in HCC progression by regulating the methylation of miR-9 CpG island. As shown, ZNFX1-AS1 inhibits the expression of DNMT1, which reduces the expression of miR-9. However, one limitation of this mechanism is that DNMT1 is a pan-DNA methyltransferase; it does not specifically regulate the expression of miR-9. Therefore, another potential target of ZNFX1 is needed to explore the functional role of ZNFX1-AS1 in HCC progression in a future study and we should further determine whether miR-9 contributes to this function.
Conclusion
Taken all together, we demonstrated that lncRNA ZNFX1-AS1 is significantly downregulated in HCC samples and cell lines, and low expression of ZNFX1-AS1 is associated with poor prognosis for HCC patients. Furthermore, we also showed that ZNFX1-AS1 suppresses HCC progression by regulating the methylation of miR-9 CpG island. LncRNA ZNFX1-AS1 may serve as a potential tumor suppressor gene.
Acknowledgments
This project was supported by National Natural Science Foundation of China (grant number 81201678); National Natural Science Foundation of China (grant number 81472845); The Key Project of Scientific Research Innovation Program of Shanghai Municipal Education Commission (grant number 14ZZ112).
Disclosure
The authors report no conflicts of interest in this work.
References
Jemal A, Bray F, Center MM, Ferlay J, Ward E, Forman D. Global cancer statistics. CA Cancer J Clin. 2011;61(2):69–90. | ||
Kiyosawa K, Umemura T, Ichijo T, et al. Hepatocellular carcinoma: recent trends in Japan. Gastroenterology. 2004;127(5Suppl 1):S17–S26. | ||
Blum HE. Hepatocellular carcinoma: therapy and prevention. World J Gastroenterol. 2005;11(47):7391–7400. | ||
Forner A, Llovet JM, Bruix J. Hepatocellular carcinoma. Lancet. 2012;379(9822):1245–1255. | ||
Budhu A, Forgues M, Ye QH, et al. Prediction of various metastases, recurrence and prognosis in hepatocellular carcinoma based on a unique immune response signature of the liver microenvironment. Cancer Cell. 2006;10(2):99–111. | ||
Ravasi T, Suzuki H, Pang KC, et al. Experimental validation of the regulated expression of large numbers of non-coding RNAs from the mouse genome. Genome Res. 2006;16(1):11–19. | ||
Kapranov P, Cheng J, Dike S, et al. RNA maps reveal new RNA classes and a possible function for pervasive transcription. Science. 2007;316(5830):1484–1488. | ||
Kapranov P, Willingham AT, Gingeras TR. Genome-wide transcription and the implications for genomic organization. Nat Rev Genet. 2007;8(6):413–423. | ||
Mercer TR, Dinger ME, Sunkin SM, Mehler MF, Mattick JS. Specific expression of long noncoding RNAs in the mouse brain. Proc Natl Acad Sci USA. 2008;105(2):716–721. | ||
Feng J, Bi C, Clark BS, Mady R, Shah P, Kohtz JD. The Evf-2 noncoding RNA is transcribed from the Dlx-5/6 ultraconserved region and functions as a Dlx-2 transcriptional coactivator. Genes Dev. 2006;20(11):1470–1484. | ||
Wang H, Iacoangeli A, Lin D, et al. Dendritic BC1 RNA in translational control mechanisms. J Cell Biol. 2005;171(5):811–821. | ||
Yan MD, Hong CC, Lai GM, Cheng AL, Lin YW, Chuang SE. Identification and characterization of a novel gene Saf transcribed from the opposite strand of Fas. Hum Mol Genet. 2005;14(11):1465–1474. | ||
Shi X, Sun M, Liu H, Yao Y, Song Y. Long non-coding RNAs: a new frontier in the study of human diseases. Cancer Lett. 2013;339(2):159–166. | ||
Gibb EA, Brown CJ, Lam WL. The functional role of long noncoding RNA in human carcinomas. Mol Cancer. 2011;10(2):38–39. | ||
Huarte M, Rinn JL. Large non-coding RNAs: missing links in cancer? Hum Mol Genet. 2010;19(R2):152–161. | ||
Korneev SA, Korneeva EI, Lagarkova MA, Kiselev SL, Critchley G, O’Shea M. Novel noncoding antisense RNA transcribed from human anti-NOS2A locus is differentially regulated during neuronal differentiation of embryonic stem cells. RNA. 2008;14(10):2030–2037. | ||
Miyoshi N, Wagatsuma H, Wakana S, et al. Identification of an imprinted gene, Meg3/Gtl2 and its human homologue MEG3, first mapped on mouse distal chromosome 12 and human chromosome14q. Genes Cells. 2000;5(3):211–220. | ||
Zhang X, Rice K, Wang Y, et al. Maternally expressed gene 3 (MEG3) noncoding ribonucleic acid: isoform structure, expression, and functions. Endocrinology. 2010;151(3):939–947. | ||
Zhang X, Zhou Y, Mehta KR, et al. A pituitary-derived MEG3 isoform functions as a growth suppressor in tumor cells. J Clin Endocrinol Metab. 2003;88(11):5119–5126. | ||
Gabory A, Jammes H, Dandolo L. The H19 locus: role of an imprinted non-coding RNA in growth and development. Bioessays. 2010;32(6):473–480. | ||
Iacoangeli A, Lin Y, Morley EJ, et al. BC200 RNA in invasive and preinvasive breast cancer. Carcinogenesis. 2004;25(11):2125–2133. | ||
Chen W, Bocker W, Brosius J, Tiedge H. Expression of neural BC200 RNA in human tumours. J Pathol. 1997;183(3):345–351. | ||
Rinn JL, Kertesz M, Wang JK, et al. Functional demarcation of active and silent chromatin domains in human HOX loci by noncoding RNAs. Cell. 2007;129(7):1311–1323. | ||
Guo F, Li Y, Liu Y, Wang J, Li Y, Li G. Inhibition of metastasis-associated lung adenocarcinoma transcript 1 in CaSki human cervical cancer cells suppresses cell proliferation and invasion. Acta Biochim Biophys Sin (Shanghai). 2010;42(3):224–229. | ||
Panzitt K, Tschernatsch MM, Guelly C, et al. Characterization of HULC, a novel gene with striking up-regulation in hepatocellular carcinoma, as noncoding RNA. Gastroenterology. 2007;132(1):330–342. | ||
Quagliata L, Matter MS, Piscuoglio S, et al. Long noncoding RNA HOTTIP/HOXA13 expression is associated with disease progression and predicts outcome in hepatocellular carcinoma patients. Hepatology. 2014;59(3):911–923. | ||
Lai MC, Yang Z, Zhou L, et al. Long non-coding RNA MALAT-1 overexpression predicts tumor recurrence of hepatocellular carcinoma after liver transplantation. Med Oncol. 2012;29(3):1810–1816. | ||
Yuan SX, Tao QF, Wang J, et al. Antisense long non-coding RNA PCNA-AS1 promotes tumor growth by regulating proliferating cell nuclear antigen in hepatocellular carcinoma. Cancer Lett. 2014;349(1):87–94. | ||
Askarian-Amiri ME, Crawford J, French JD, et al. SNORD-host RNA Zfas1 is a regulator of mammary development and a potential marker for breast cancer. RNA. 2011;17(5):878–891. | ||
Chen H, Xin Y, Zhou L, et al. Cisplatin and paclitaxel target significant long noncoding RNAs in laryngeal squamous cell carcinoma. Med Oncol. 2014;31(11):246–259. | ||
Rust S, Funke H, Assmann G. Mutagenically separated PCR (MS-PCR): a highly specific one step procedure for easy mutation detection. Nucleic Acids Res. 1993;21(16):3623–3629. | ||
Zhang N, Su Y, Xu L. Targeting PKCε by miR-143 regulates cell apoptosis in lung cancer. FEBS Lett. 2013;587(22):3661–3667. | ||
Cai L, Cai X. Up-regulation of miR-9 expression predicate advanced clinicopathological features and poor prognosis in patients with hepatocellular carcinoma. Diagn Pathol. 2014;9:1000–1006. | ||
Bruix J, Gores GJ, Mazzaferro V. Hepatocellular carcinoma: clinical frontiers and perspectives. Gut. 2014;63(5):844–855. | ||
Yuan JH, Yang F, Wang F, et al. A long noncoding RNA activated by TGF-beta promotes the invasion-metastasis cascade in hepatocellular carcinoma. Cancer Cell. 2014;25(5):666–681. | ||
Tanner MM, Tirkkonen M, Kallioniemi A, et al. Increased copy number at 20ql3 in breast cancer: defining the critical region and exclusion of candidate genes. Cancer Res. 1994;54(16):4257–4260. | ||
Bar-Shira A, Pinthus JH, Rozovsky U, et al. Multiple genes in human 20q13 chromosomal region are involved in an advanced prostate cancer xenograft. Cancer Res. 2002;62(23):6803–6807. |
Supplementary materials
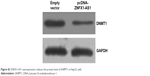  | Figure S3 ZNFX1-AS1 overexpression reduces the protein level of DNMT1 in HepG2 cells. |
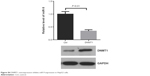  | Figure S4 DNMT1 overexpression inhibits miR-9 expression in HepG2 cells. |
 © 2016 The Author(s). This work is published and licensed by Dove Medical Press Limited. The full terms of this license are available at https://www.dovepress.com/terms.php and incorporate the Creative Commons Attribution - Non Commercial (unported, v3.0) License.
By accessing the work you hereby accept the Terms. Non-commercial uses of the work are permitted without any further permission from Dove Medical Press Limited, provided the work is properly attributed. For permission for commercial use of this work, please see paragraphs 4.2 and 5 of our Terms.
© 2016 The Author(s). This work is published and licensed by Dove Medical Press Limited. The full terms of this license are available at https://www.dovepress.com/terms.php and incorporate the Creative Commons Attribution - Non Commercial (unported, v3.0) License.
By accessing the work you hereby accept the Terms. Non-commercial uses of the work are permitted without any further permission from Dove Medical Press Limited, provided the work is properly attributed. For permission for commercial use of this work, please see paragraphs 4.2 and 5 of our Terms.