Back to Journals » OncoTargets and Therapy » Volume 12
Long Noncoding RNA SNHG16 Sponges miR-182-5p And miR-128-3p To Promote Retinoblastoma Cell Migration And Invasion By Targeting LASP1
Authors Yang L, Zhang L, Lu L, Wang Y
Received 15 April 2019
Accepted for publication 21 August 2019
Published 18 October 2019 Volume 2019:12 Pages 8653—8662
DOI https://doi.org/10.2147/OTT.S212352
Checked for plagiarism Yes
Review by Single anonymous peer review
Peer reviewer comments 2
Editor who approved publication: Prof. Dr. Takuya Aoki
Lidong Yang, Liyou Zhang, Lu Lu, Yan Wang
Department of Ocular Fundus Disease, Cangzhou Eye Hospital, Cangzhou Central Hospital, Cangzhou 061001, People’s Republic of China
Correspondence: Lidong Yang
Department of Ocular Fundus Disease, Cangzhou Eye Hospital, Cangzhou Central Hospital, #7-1 South Fuyang Avenue, Cangzhou 061001, People’s Republic of China
Email [email protected]
Background: Long noncoding RNAs (lncRNAs) are dysregulated and play an important role in the tumorigenesis and progression of cancers. However, the potential roles of SNHG16 in retinoblastoma progression still remain largely unclear.
Materials and methods: The expression levels of SNHG16, miR-182-5p, miR-128-3p and LASP1 in retinoblastoma tissues and cell lines were detected using qRT-PCR. The migratory and invasive abilities of retinoblastoma cells were assessed using Transwell assay. The regulatory relationships among SNHG16, miR-182-5p, miR-128-3p and LASP1 were analyzed through bioinformatics prediction and validated by luciferase reporter assay and Western blot.
Results: Here, we demonstrated that SNHG16 was frequently up-regulated in retinoblastoma tissue samples and cell lines. Clinicopathological features showed that high levels of SNHG16 were significantly associated with retinoblastoma metastasis and predicted poor overall survival. Functional studies demonstrated that knockdown of SNHG16 suppressed retinoblastoma cell migration and invasion. Mechanistic investigation revealed that SNHG16 exerted its oncogenic activity through increasing LASP1 expression and sponging miR-182-5p and miR-128-3p.
Conclusion: Taken together, our findings suggest SNHG16 as a novel biomarker and therapeutic target against retinoblastoma.
Keywords: long noncoding RNA, retinoblastoma, SNHG16, metastasis, LASP1
Introduction
Retinoblastoma is the most common childhood intraocular malignancy and approximately 8000 cases are diagnosed yearly worldwide.1 In United States, there were 3540 new estimated cases and 350 deaths with eye and orbit cancer in 2018.2 Choroidal invasion in retinoblastoma eyes is commonly observed pathologically. Tumor invasion was initiated from the retina to the sclera and post-laminar optic nerve and even metastasize to the central nervous system for late-stage retinoblastoma.3 Once diagnosed with metastasis, the overall survival of patients was significantly deteriorated. Hence, it is urgent to understand the molecular mechanisms and develop novel therapeutic strategies for retinoblastoma treatment.
Long noncoding RNAs (lncRNAs) are dysregulated and act as key regulators of several hallmarks of cancer biology, including cell growth, metastasis and drug resistance.4–6 LncRNAs could function as competing endogenous RNA (ceRNA) or “RNA sponges” of miRNAs and reduce their regulatory effect on target mRNA and target mRNA-mediated biological processes.7 Recent advancements in surveying lncRNA functions suggest that lncRNAs hold strong promise as novel biomarkers and therapeutic targets for cancer.
SNHG16 (small nucleolar RNA host gene 16) is a processed long noncoding RNA with a length of 7585 nucleotides and located on chromosome 17q25.1. SNHG16 was first reported to be up-regulated in colorectal cancer and silencing of SNHG16 suppressed cell viability and migration and increased apoptotic cell death.8 In esophageal squamous cell carcinoma, high SNHG16 expression level is an independent predictor of poor survival and promotes cell proliferation and invasion by regulating Wnt/β-catenin signaling pathway.9 These findings suggest an oncogene role of SNHG16 in certain tumors. Accumulating evidence demonstrate that SNHG16 can act as ceRNA to regulate target gene expression through sponging several miRNAs, including miR-520d-3p, miR-135a, miR-195, miR-146a-5p and miR-373.10–14 However, the potential functions of SNHG16 in retinoblastoma are still largely unknown.
The purpose of this study was to investigate the expression profile, clinical significance and functional roles of SNHG16 in retinoblastoma and to also explore the underlying mechanism in which SNHG16 affects the progression of retinoblastoma. We found that SNHG16 was commonly up-regulated in retinoblastoma tissues and cell lines and significantly associated with retinoblastoma metastasis. Further studies revealed that SNHG16 promoted migration and invasion of retinoblastoma cells through sponging miR-182-5p and miR-128-3p.
Materials And Methods
Patients And Samples
Seventy-six human retinoblastoma specimens (42 males and 34 females, aged from 5 months to 15.2 years and averaged 4.8 years) and 15 normal retina tissues (9 males and 6 females, aged from 4.4 to 14.6 years and averaged 6.7 years) were collected from Cangzhou Central Hospital between 2013 and 2017. According to the international classification of retinoblastoma (ICRB), stages I–V of retinoblastoma patients were 8, 7, 36, 18 and 7, respectively. As most retinoblastoma patients were infants, radiotherapy and/or chemotherapy will cause serious physical injury. The routine treatment for retinoblastoma patients is enucleation. Tumor samples were collected during enucleation surgery and normal retina tissues were mainly obtained from the donation of accidental death. None of the subjects had received any preoperative radiotherapy and/or chemotherapy. Histological and pathological diagnoses of these specimens were confirmed and classified by two experienced clinical pathologists. Median follow-up time was 18.0 months (range 1.0–58.0 months). Written informed consent was obtained from all patients and the study was carried out in accordance with Declaration of Helsinki and approved by the Ethics Committee of Cangzhou Central Hospital (No. 2017-069).
Cell Culture And Transfection
Human retinoblastoma cell lines WERI-RB1, SO-RB50, Y79 and human retinal pigment epithelial cell line ARPE-19 were purchased from the American Type Culture Collection (ATCC) and maintained in RPMI-1640 medium (Gibco) supplemented with 10% fetal bovine serum (FBS, Hyclone) under a humidified atmosphere at 37 °C with 5% CO2. 293T cells were purchased from the Chinese Academy of Sciences Cell Bank (Shanghai, China) and cultured in Dulbecco’s Modified Eagle’s Medium (Gibco). SNHG16-specific siRNA (si-SNHG16), siRNA negative control (si-NC), miR-182-5p mimics, miR-128-3p mimics, miR-182-5p inhibitor (miR-182-5p in), miR-128-3p inhibitor (miR-128-3p in) and miRNA negative control (miRNA NC) were synthesized by GenePharma (GenePharma, Shanghai, China). The transfection was performed using Lipofectamine 2000 (Invitrogen, USA) according to the manufacturer’s instructions.
Cell Migration And Invasion Assays
Cell migration and invasion assays were determined through transwell chamber (8 μM pore size, (BD Biosciences, Bedford, MA, USA). For migration assay, cells were plated into the upper chamber. For invasion assay, cells were added into the upper chamber pre-coated with 50 μL Matrigel (BD Biosciences) and cultured with serum-free medium. The lower chamber was filled with RPMI-1640 medium containing 20% FBS. After 24 hrs, the invaded cells were fixed and stained with 1% crystal violet. The migrated and invaded cells were counted under a microscope (Olympus, Tokyo, Japan).
RNA Isolation And qRT-PCR
Total RNA from the retinoblastoma tissues and cell lines was extracted using TRIzol reagent (Ambion, Life Technologies). Total RNA was reverse-transcribed with the PrimeScript RT reagent kit (Promega, Madison, WI, USA). Determinations of SNHG16, mRNA and miRNAs were performed with the SYBR Premix ExTaq (TaKaRa) and Mir-X miRNA First Strand Synthesis Kit (TaKaRa), respectively. β-actin and U6 were used as the normalization controls for SNHG16, mRNA and miRNAs, respectively. The relative expression levels were calculated using 2−ΔΔCT method.
Western Blot Analysis
Cells were lysed in RIPA buffer. Equal amounts of protein samples were separated using 10% SDS-PAGE gels and transferred to PVDF membranes (Millipore). The membranes were incubated with primary antibodies (E-cadherin, N-cadherin, vimentin, LASP1 and β-actin) (1:1000; Cell Signaling Technology) at 4 °C overnight, respectively. Then, the membranes were incubated with secondary antibodies at room temperature for 2 hrs. Protein bands were detected with chemiluminescence, and the protein expression levels were normalized to β-actin.
Immunohistochemical Staining
Tissues were formalin-fixed, paraffin-embedded (4 μm), and the levels of LASP1 (1:250; Cell Signaling Technology) were analyzed following the standard protocol. Stained tissue sections were evaluated separately by two pathologists.
Luciferase Reporter Assay
A fragment of the wild and mutant reporter plasmids of SNHG16 (WT-182-SNHG16, Mut-182-SNHG16, WT-128-SNHG16, Mut-128-SNHG16) and LASP1 (WT-182-LASP1, Mut-182-LASP1, WT-128-LASP1, Mut-128-LASP1) containing the putative binding sites for miR-182-5p or miR-128-3p were synthesized by GenePharma (GenePharma) and cloned into psiCHECK-2 vectors (Invitrogen). For luciferase assay, cells were co-transfected WT or Mut reporter with miRNA mimics or miRNA NC. After 48-hr transfection, luciferase activity was determined using a Dual Luciferase Reporter Assay Kit (Promega, Madison, WI, USA).
Statistical Analysis
All data were expressed as the mean ± standard deviation (SD) and each experiment was performed in triplicate at least. The differences between groups were analyzed by two-tailed Student’s t-test or one-way ANOVA using Graphpad Prism 5 software (GraphPad, San Diego, CA, USA). A p value < 0.05 was considered statistically significant.
Results
SNHG16 Is Significantly Up-Regulated In Retinoblastoma And Associated With Poor Prognosis
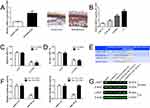
To investigate the functional roles of SNHG16 in the development of retinoblastoma, the expression levels of SNHG16 in 76 retinoblastoma tissues and normal retina tissues were determined using qRT-PCR. The results showed that SNHG16 was elevated in most retinoblastoma tissues (58/76, 76.31%) when compared with non-tumor tissues (Figure 1A and B). Additionally, clinicopathological feature analysis results demonstrated that up-regulated SNHG16 expression was positively correlated with TNM stage (p=0.004), choroidal invasion (p=0.011) and optic nerve invasion (p=0.009; Table 1). Kaplan–Meier analysis and log rank test revealed that elevated SNHG16 level was markedly associated with poor overall survival of retinoblastoma patients (p=0.006; Figure 1C). SNHG16 expression was also up-regulated in retinoblastoma cell lines compared with normal retina cell lines (Figure 1D). These results indicated that SNHG16 may play an important role in retinoblastoma metastasis.
 |
Table 1 SNHG16 Expression And Clinicopathologic Features Of Retinoblastoma Patients |
Knockdown Of SNHG16 Inhibits Migration And Invasion Of SO-RB50 And Y79 Cells
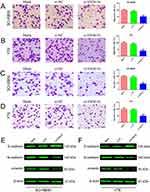
Next, we determined whether SNHG16 could regulate cell migration and invasion of retinoblastoma cells. The specific siRNA of SNHG16 significantly decreased the expression level of SNHG16 in SO-RB50 and Y79 cells (Supplementary Figure 1). The migratory and invasive abilities of retinoblastoma cells were assessed using Transwell migration and invasion assays. We observed strong inhibition of cellular migration and invasion following SNHG16 knockdown relative to control cells (Figure 2A–D). In addition, knockdown of SNHG16 increased E-cadherin expression and decreased N-cadherin and vimentin expression in both SO-RB50 and Y79 cells (Figure 2E and F). These findings suggested that SNHG16 promotes migration and invasion of retinoblastoma cells.
SNHG16 Directly Binds To miR-182-5p And miR-128-3p
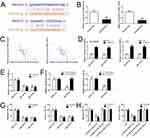
Given that lncRNAs could bind to different miRNAs and regulate downstream genes, we found that the 3ʹUTR of SNHG16 contains binding sites of miR-182-5p and miR-128-3p predicted through starBase (http://starbase.sysu.edu.cn/) (Figure 3A). miR-182-5p and miR-128-3p expression levels in retinoblastoma tissues were determined using qRT-PCR. The results showed a down-regulation of miR-182-5p and miR-128-3p in retinoblastoma (Figure 3B). Pearson correlation analysis revealed that SNHG16 expression was negatively correlated with miR-182-5p and miR-128-3p in retinoblastoma (Figure 3C). Knockdown of SNHG16 increased miR-182-5p and miR-128-3p expression in retinoblastoma cells (Figure 3D). Conversely, transfection with miR-182-5p and miR-128-3p mimics also decreased SNHG16 expression (Figure 3E). We further validated the target relationship between SNHG16 and miR-182-5p or miR-128-3p using luciferase reporter assay. Co-transfection miR-182-5p or miR-128-3p mimics with SNHG16 3ʹUTR WT vector significantly decreased luciferase activity compared with miRNA negative control or SNHG16 3ʹUTR Mut vector (Figure 3F). These results indicate that SNHG16 directly interacts with miR-182-5p and miR-128-3p.
To explore whether miR-182-5p and miR-128-3p could regulate retinoblastoma metastasis, SO-RB50 and Y79 cells were transfected with miR-182-5p and miR-128-3p mimics or miRNA negative control and the relative expressions of miR-182-5p and miR-128-3p were determined using qRT-PCR (Supplementary Figure 2). Inhibited cell migrations were observed in SO-RB50 and Y79 cells induced by overexpression of miR-182-5p or miR-128-3p (Figure 3G). Consistent with this observation, transfection with miR-182-5p and miR-128-3p mimics also decreased the number of cells to invade through a layer of extracellular matrix. Additionally, miR-182-5p or miR-128-3p inhibitor attenuated the effects of SNHG16 on migration and invasion in SO-RB50 and Y79 cells (Figure 3H). The results suggest that miR-182-5p and miR-128-3p function as a tumor suppressor and mediate SNHG16-induced metastasis in the progression of retinoblastoma.
SNHG16 Promotes The Migration And Invasion Of Retinoblastoma By Sponging miR-182-5p And miR-128-3p To Regulate LASP1
According to previous studies and our bioinformatics analysis, we found that LASP1 was closely related to the migration and invasion of retinoblastoma cells. LASP1 was up-regulated in both retinoblastoma cells and tissues (Figure 4A and B) and knockdown of LASP1 inhibited migration and invasion of retinoblastoma cells (Figure 4C and D). As shown in Figure 4E, the 3ʹUTR of LASP1 was predicted to sponge to miR-182-5p and miR-128-3p by TargetScan, starBase and microRNA.org. To verify these predicted relationships, luciferase reporter assay was performed. The luciferase activity of LASP1 3ʹUTR WT reporter vector, but not Mut vector, was obviously reduced after transfected with miR-182-5p or miR-128-3p mimics compared with miRNA negative control (Figure 4F). Western blot result demonstrated that knockdown of SNHG16 decreased LASP1 expression level, but LASP1 was increased following by transfection with miR-182-5p or miR-128-3p inhibitor compared with miRNA NC or si-SNHG16 group (Figure 4G). Taken together, the results indicate that SNHG16 decoyed miR-182-5p or miR-128-3p to facilitate LASP1-mediated migration and invasion in retinoblastoma.
Discussion
SNHG16 was frequently up-regulated and has been identified as oncogene in most human cancers, yet its function in the pathogenesis of retinoblastoma is unclear. In this study, we found that SNHG16 was significantly highly expressed in retinoblastoma tissues and cell lines compared with normal controls. We also observed that a high level of SNHG16 was associated with poor prognosis of patients with retinoblastoma. Clinicopathological features further revealed that up-regulated SNHG16 expression was positively correlated with TNM stage, present choroidal invasion and present optic nerve invasion. These findings indicated that SNHG16 may exert its oncogene role through promoting cell metastasis in the progression of retinoblastoma.
Tumor metastasis causes more than 90% of cancer cell mortality. Consistent with the previous reports, we observed that transfection with specific siRNA for SNHG16 obviously decreased the migratory and invasive abilities of retinoblastoma cells compared with negative control. Epithelial–mesenchymal transition (EMT) is a crucial process responsible for cancer metastasis.15–17 Knockdown of SNHG16 also regulates the expression of E-cadherin, N-cadherin and vimentin. These results suggest an important role of SNHG16 in the retinoblastoma metastasis.
We identified that miR-182-5p and miR-128-3p were down-regulated in retinoblastoma, negatively correlated with SNHG16 expression. Overexpression of miR-182-5p or miR-128-3p inhibited cell migration and invasion of retinoblastoma cells. Further investigation illuminated that miR-182-5p and miR-128-3p interacted and regulated SNHG16 expression. In the previous studies, miR-182-5p was reported to function as a tumor suppressor of cancer metastasis through regulating RAB27A, FOXO3a, ARRDC3, PTEN and Cofilin 1.18–21 Dysregulated miR-182-5p was also involved in nonalcoholic steatohepatitis, chondrogenesis, lupus nephritis and cerebral ischemia-reperfusion injury.23–26 Several studies also indicated that miR-128-3p was down-regulated in several types of cancer, and restoration of miR-128-3p inhibited cell migration and invasion. For example, miR-128-3p inhibits metastasis and EMT by targeting ZEB1 in esophageal squamous cell cancer, reduces the migration and invasion ability of glioma cell through regulating GREM1 and suppresses osteosarcoma proliferation and metastasis by reducing VEGFC.27–29 miR-128-3p also plays a key role in regulating the development and progression of rheumatoid arthritis, type 2 diabetes and atrial fibrillation.30–32
LIM and SH3 protein 1 (LASP1) was located at 17q12 and initially identified in breast cancer metastatic lymph nodes.33 Recent studies have suggested that LASP1 is an important prognostic marker and associated with tumor size, stage and metastasis in different tumors.34 LASP1 has been identified as the target gene of many miRNAs regulating metastasis, such as miR-133ab, miR-29ab, miR-1, miR-203 and miR-218.35–41 In this study, we observed that SNHG16 promoted migration and invasion of retinoblastoma cells by regulating LASP1 via competitively sponging miR-182-5p and miR-128-3p. LASP1 promoting retinoblastoma metastasis may be through its interaction with cytoskeleton and increased nuclear translocation of CXCR1-4, UHRF1 and AP-1.42–44 However, the detailed mechanisms still need to be further investigated.
Conclusion
In summary, we identified SNHG16 as a metastasis-specific oncogenic lncRNA in retinoblastoma and revealed a novel ceRNA regulatory pathway in which SNHG16 increased LASP1 expression by sponging miR-182-5p and miR-128-3p. Thus, SNHG16 is a novel biomarker and therapeutic potential target against retinoblastoma.
Disclosure
The authors declare no competing financial interests in this work.
Reference
1. Pascual-Pasto G, Bazan-Peregrino M, Olaciregui NG, et al. Therapeutic targeting of the RB1 pathway in retinoblastoma with the oncolytic adenovirus VCN-01. Sci Transl Med. 2019;11(476):
2. Siegel RL, Miller KD, Jemal A. Cancer statistics, 2018. CA Cancer J Clin. 2018;68(1):7–30. doi:10.3322/caac.21442
3. Gündüz K, Müftüoglu O, Günalp I, Unal E, Taçyildiz N. Metastatic retinoblastoma clinical features, treatment, and prognosis. Ophthalmology. 2006;113(9):1558–1566. doi:10.1016/j.ophtha.2006.03.039
4. Bhan A, Soleimani M, Mandal SS. Long noncoding RNA and cancer: a new paradigm. Cancer Res. 2017;77(15):3965–3981. doi:10.1158/0008-5472.CAN-16-2634
5. Bach DH, Lee SK. Long noncoding RNAs in cancer cells. Cancer Lett. 2018;419:152–166. doi:10.1016/j.canlet.2018.01.053
6. Xie W, Yuan S, Sun Z, Li Y. Long noncoding and circular RNAs in lung cancer: advances and perspectives. Epigenomics. 2016;8(9):1275–1287. doi:10.2217/epi-2016-0036
7. Tay Y, Rinn J, Pandolfi PP. The multilayered complexity of ceRNA crosstalk and competition. Nature. 2014;505(7483):344–352. doi:10.1038/nature12986
8. Christensen LL, True K, Hamilton MP, et al. SNHG16 is regulated by the Wnt pathway in colorectal cancer and affects genes involved in lipid metabolism. Mol Oncol. 2016;10(8):1266–1282. doi:10.1016/j.molonc.2016.06.003
9. Han GH, Lu KJ, Wang P, Ye J, Ye -Y-Y, Huang J-X. LncRNA SNHG16 predicts poor prognosis in ESCC and promotes cell proliferation and invasion by regulating Wnt/β-catenin signaling pathway. Eur Rev Med Pharmacol Sci. 2018;22(12):3795–3803. doi:10.26355/eurrev_201806_15262
10. Zhao W, Fu H, Zhang S, et al. LncRNA SNHG16 drives proliferation, migration, and invasion of hemangioma endothelial cell through modulation of miR-520d-3p/STAT3 axis. Cancer Med. 2018;7:3311–3320.
11. Wang X, Kan J, Han J, Zhang W, Bai L, Wu H. LncRNA SNHG16 functions as an oncogene by sponging MiR-135a and promotes JAK2/STAT3 signal pathway in gastric cancer. J Cancer. 2019;10(4):1013–1022. doi:10.7150/jca.29527
12. Xie X, Xu X, Sun C, Yu Z. Long intergenic noncoding RNA SNHG16 interacts with miR-195 to promote proliferation, invasion and tumorigenesis in hepatocellular carcinoma. Exp Cell Res. 2019;383:111501. doi:10.1016/j.yexcr.2019.111501
13. Zhou Z, Zhu Y, Gao G, Zhang Y. Long noncoding RNA SNHG16 targets miR-146a-5p/CCL5 to regulate LPS-induced WI-38 cell apoptosis and inflammation in acute pneumonia. Life Sci. 2019;228:189–197. doi:10.1016/j.lfs.2019.05.008
14. Zhou XY, Liu H, Ding ZB, et al. lncRNA SNHG16 promotes glioma tumorigenicity through miR-373/EGFR axis by activating PI3K/AKT pathway. Genomics. 2019;
15. Voulgari A, Pintzas A. Epithelial-mesenchymal transition in cancer metastasis: mechanisms, markers and strategies to overcome drug resistance in the clinic. Biochim Biophys Acta. 2009;1796(2):75–90. doi:10.1016/j.bbcan.2009.03.002
16. Chaffer CL, San Juan BP, Lim E, Weinberg RA. EMT, cell plasticity and metastasis. Cancer Metastasis Rev. 2016;35(4):645–654. doi:10.1007/s10555-016-9648-7
17. Ombrato L, Malanchi I. The EMT universe: space between cancer cell dissemination and metastasis initiation. Crit Rev Oncog. 2014;19(5):349–361.
18. Li Y, Chen S, Shan Z, et al. miR-182-5p improves the viability, mitosis, migration, and invasion ability of human gastric cancer cells by down-regulating RAB27A. Biosci Rep. 2017;37(3):
19. Cao MQ, You AB, Zhu XD, et al. miR-182-5p promotes hepatocellular carcinoma progression by repressing FOXO3a. J Hematol Oncol. 2018;11(1):12. doi:10.1186/s13045-018-0555-y
20. Yao J, Xu C, Fang Z, et al. Androgen receptor regulated microRNA miR-182-5p promotes prostate cancer progression by targeting the ARRDC3/ITGB4 pathway. Biochem Biophys Res Commun. 2016;474(1):213–219. doi:10.1016/j.bbrc.2016.04.107
21. Zhao YS, Yang WC, Xin HW, Han JX, Ma SG. miR-182-5p knockdown targeting PTEN inhibits cell proliferation and invasion of breast cancer cells. Yonsei Med J. 2019;60(2):148–157. doi:10.3349/ymj.2019.60.2.148
22. Wang F, Wu D, Xu Z, et al. miR-182-5p affects human bladder cancer cell proliferation, migration and invasion through regulating Cofilin 1. Cancer Cell Int. 2019;19:42. doi:10.1186/s12935-019-0758-5
23. Liang Q, Chen H, Xu X, Jiang W. miR-182-5p attenuates high-fat-diet-induced nonalcoholic steatohepatitis in mice. Ann Hepatol. 2018;18(1):116–125. doi:10.5604/01.3001.0012.7902
24. Bai M, Yin H, Zhao J, Li Y, Wu Y. miR-182-5p overexpression inhibits chondrogenesis by down-regulating PTHLH. Cell Biol Int. 2019;43(3):222–232. doi:10.1002/cbin.11047
25. Wang X, Wang G, Zhang X, et al. Inhibition of microRNA-182-5p contributes to attenuation of lupus nephritis via Foxo1 signaling. Exp Cell Res. 2018;373(1–2):91–98. doi:10.1016/j.yexcr.2018.09.026
26. Wang J, Xu Z, Chen X, et al. MicroRNA-182-5p attenuates cerebral ischemia-reperfusion injury by targeting Toll-like receptor 4. Biochem Biophys Res Commun. 2018;505(3):677–684. doi:10.1016/j.bbrc.2018.09.165
27. Zhao L, Li R, Xu S, et al. Tumor suppressor miR-128-3p inhibits metastasis and epithelial-mesenchymal transition by targeting ZEB1 in esophageal squamous-cell cancer. Acta Biochim Biophys Sin (Shanghai). 2018;50(2):171–180. doi:10.1093/abbs/gmx132
28. Fu C, Li D, Zhang X, Liu N, Chi G, Jin X. LncRNA PVT1 facilitates tumorigenesis and progression of glioma via regulation of MiR-128-3p/GREM1 axis and BMP signaling pathway. Neurotherapeutics. 2018;15(4):1139–1157. doi:10.1007/s13311-018-0649-9
29. Zhang C, Xie L, Liang H, Cui Y. LncRNA MIAT facilitates osteosarcoma progression by regulating miR-128-3p/VEGFC axis. IUBMB Life. 2019;71:845–853. doi:10.1002/iub.v71.7
30. Xia Z, Meng F, Liu Y, et al. Decreased MiR-128-3p alleviates the progression of rheumatoid arthritis by up-regulating the expression of TNFAIP3. Biosci Rep. 2018;38(4):
31. Wang XY, Zhang XZ, Li F, Ji Q-R. MiR-128-3p accelerates cardiovascular calcification and insulin resistance through ISL1-dependent Wnt pathway in type 2 diabetes mellitus rats. J Cell Physiol. 2019;234(4):4997–5010. doi:10.1002/jcp.27300
32. Cao F, Li Z, Ding WM, Yan L, Zhao Q-Y. LncRNA PVT1 regulates atrial fibrosis via miR-128-3p-SP1-TGF-β1-Smad axis in atrial fibrillation. Mol Med. 2019;25(1):7. doi:10.1186/s10020-019-0074-5
33. Tomasetto C, Moog-Lutz C, Régnier CH, Schreiber V, Basset P, Rio MC. Lasp-1 (MLN 50) defines a new LIM protein subfamily characterized by the association of LIM and SH3 domains. FEBS Lett. 1995;373(3):245–249. doi:10.1016/0014-5793(95)01040-l
34. Hu Z, Wang X, Cui Y, Li C, Wang S. LASP1 in tumor and tumor microenvironment. Curr Mol Med. 2017;17(8):541–548. doi:10.2174/1566524018666180222115103
35. Wang H, An H, Wang B, et al. miR-133a represses tumour growth and metastasis in colorectal cancer by targeting LIM and SH3 protein 1 and inhibiting the MAPK pathway. Eur J Cancer. 2013;49(18):3924–3935. doi:10.1016/j.ejca.2013.07.149
36. Li H, Xiang Z, Liu Y, Xu B, Tang J. MicroRNA-133b inhibits proliferation, cellular migration, and invasion via targeting LASP1 in hepatocarcinoma cells. Oncol Res. 2017;25(8):1269–1282. doi:10.3727/096504017X14850151453092
37. Hu Z, Cui Y, Zhou Y, et al. MicroRNA-29a plays a suppressive role in non-small cell lung cancer cells via targeting LASP1. Onco Targets Ther. 2016;9:6999–7009. doi:10.2147/OTT.S116509
38. Li H, Liu G, Pan K, Miao X, Xie Y. Methylation-induced downregulation and tumor suppressive role of microRNA-29b in gastric cancer through targeting LASP1. Oncotarget. 2017;8(56):95880–95895. doi:10.18632/oncotarget.21431
39. Du YY, Zhao LM, Chen L, et al. The tumor-suppressive function of miR-1 by targeting LASP1 and TAGLN2 in esophageal squamous cell carcinoma. J Gastroenterol Hepatol. 2016;31(2):384–393. doi:10.1111/jgh.13180
40. Tan J, Jing YY, Han L, et al. MicroRNA-203 inhibits invasion and induces apoptosis of laryngeal cancer cells via targeting LASP1. Eur Rev Med Pharmacol Sci. 2018;22(19):6350–6357. doi:10.26355/eurrev_201810_16046
41. Nishikawa R, Goto Y, Sakamoto S, et al. Tumor-suppressive microRNA-218 inhibits cancer cell migration and invasion via targeting of LASP1 in prostate cancer. Cancer Sci. 2014;105(7):802–811. doi:10.1111/cas.12441
42. Raman D, Sai J, Neel NF, Chew CS, Richmond A, Unutmaz D. LIM and SH3 protein-1 modulates CXCR2-mediated cell migration. PLoS One. 2010;5(4):e10050. doi:10.1371/journal.pone.0010050
43. Duvall-Noelle N, Karwandyar A, Richmond A, Raman D. LASP-1: a nuclear hub for the UHRF1-DNMT1-G9a-Snail1 complex. Oncogene. 2016;35(9):1122–1133. doi:10.1038/onc.2015.166
44. Endres M, Kneitz S, Orth MF, Perera RK, Zernecke A, Butt E. Regulation of matrix metalloproteinases (MMPs) expression and secretion in MDA-MB-231 breast cancer cells by LIM and SH3 protein 1 (LASP1). Oncotarget. 2016;7(39):64244–64259. doi:10.18632/oncotarget.11720
 © 2019 The Author(s). This work is published and licensed by Dove Medical Press Limited. The full terms of this license are available at https://www.dovepress.com/terms.php and incorporate the Creative Commons Attribution - Non Commercial (unported, v3.0) License.
By accessing the work you hereby accept the Terms. Non-commercial uses of the work are permitted without any further permission from Dove Medical Press Limited, provided the work is properly attributed. For permission for commercial use of this work, please see paragraphs 4.2 and 5 of our Terms.
© 2019 The Author(s). This work is published and licensed by Dove Medical Press Limited. The full terms of this license are available at https://www.dovepress.com/terms.php and incorporate the Creative Commons Attribution - Non Commercial (unported, v3.0) License.
By accessing the work you hereby accept the Terms. Non-commercial uses of the work are permitted without any further permission from Dove Medical Press Limited, provided the work is properly attributed. For permission for commercial use of this work, please see paragraphs 4.2 and 5 of our Terms.