Back to Journals » OncoTargets and Therapy » Volume 13
Long Noncoding RNA SNHG12 Indicates the Prognosis and Accelerates Tumorigenesis of Diffuse Large B-Cell Lymphoma Through Sponging microR-195
Authors Chen LY, Zhang XM, Han BQ, Dai HB
Received 12 February 2020
Accepted for publication 11 May 2020
Published 12 June 2020 Volume 2020:13 Pages 5563—5574
DOI https://doi.org/10.2147/OTT.S249429
Checked for plagiarism Yes
Review by Single anonymous peer review
Peer reviewer comments 2
Editor who approved publication: Dr Sanjay Singh
Li-Yan Chen, Xiao-Min Zhang, Bi-Qing Han, Hai-Bin Dai
Department of Hematology, The 2nd Affiliated Hospital of Harbin Medical University, Harbin 150001, Heilongjiang, People’s Republic of China
Correspondence: Hai-Bin Dai
Department of Hematology, The 2nd Affiliated Hospital of Harbin Medical University, Harbin 150001, Heilongjiang, People’s Republic of China
Email [email protected]
Background: Small nucleolar RNA host gene 12 (SNHG12) expression is associated with multiple cancers, including renal cell carcinoma, prostate cancer, cervical cancer, nasopharyngeal carcinoma, colorectal cancer, and hepatocellular carcinoma. However, SNHG12 biological function is unclear in diffuse large B-cell lymphoma (DLBCL).
Methods: SNHG12 expression and associated clinicopathological characteristics were evaluated in DLBCL tissues. CCK-8 and transwell assay were used to analyze the in vitro role of SNHG12 in DLBCL progression. The xenograft model was used to explore the in vivo role of SNHG12 in DLBCL growth. The physical interaction between SNHG12 and miR-195 was confirmed using bioinformatics analysis and a dual luciferase assay.
Results: SNHG12 expression was upregulated in DLBCL tissues and correlated with patients’ prognosis. SNHG12 downregulation inhibited cell growth, migration, and invasion of DLBCL cells in vitro, while its overexpression promoted these cellular processes. Moreover, SNHG12 knockdown repressed tumorigenesis of DLBCL cells in vivo. Further experiments demonstrated that miR-195 is a target of SNHG12 in DLBCL and that their expression negatively correlates in DLBCL. SNHG12 functioned as a competing endogenous RNA for miR-195 in DLBCL cells and miR-195 upregulation abolished the effects of SNHG12 on of DLBCL progression.
Conclusion: SNHG12 predicts poor clinical outcome and serves as a novel oncogene in DLBCL via miR-195 sponging. We also suggest that SNHG12 can be used as a potential therapeutic candidate for DLBCL patients.
Keywords: diffuse large B-cell lymphoma, SNHG12, microR-195, progression, prognosis
Introduction
Lymphoma is a common cancer that develops in the hematologic and lymphoid systems, such as the lymph nodes, spleen, thymus and extra-nodal lymphatic tissues and organs.1,2 Diffuse large B-cell lymphoma (DLBCL) is one of the most prevalent types of non-Hodgkin’s lymphoma, that has an increasing incidence, accounting for almost 30% and 45.8% of non-Hodgkin’s lymphoma in Western countries and in China, respectively.3,4 DLBCL is a heterogeneous clinical, pathological and molecular entity, with a 5-year survival percentages ranging between 32 and 81%.5 Despite the optimal therapeutic response rate of the combination rituximab with chemotherapy, a significant proportion of DLBCL shows strong refractory and relapse tendencies, that suggest a limited response to first-line treatment.6,7 Currently, emerging treatment strategies based on novel molecular targets are of vital importance in improving DLBCL patients’ prognosis.
Long noncoding RNAs (lncRNAs) are transcripts of more than 200 nucleotides in length with limited or no ability to encode proteins. Increasing evidence indicated that lncRNAs can influence several biological processes, including inflammation, differentiation, proliferation, apoptosis, invasion and metastasis.8,9 Moreover, several lncRNAs have been previously reported to be involved in various cancers, including DLBCL.10 During tumorigenesis, LncRNAs that possess microRNA binding sites, can compete for shared miRNAs and interact and regulate each other’s expression levels, thus acting as competing endogenous RNAs (ceRNAs).11 The small nucleolar RNA host gene 12 (SNHG12) is located at chromosome 1p35.3 and is involved in multiple cancers, including cervical cancer, renal cell carcinoma, prostate cancer, nasopharyngeal carcinoma, colorectal cancer and hepatocellular carcinoma.12–17 However, SNHG12 biological role and related mechanisms in DLBCL remain unknown.
In the present study, we found that SNHG12 is upregulated in human DLBCL tissues and that its expression was closely associated with prognosis and clinicopathologic parameters. We showed that SNHG12 could promote the growth, migration, and invasion abilities of DLBCL cells in vitro, but also their growth in vivo. Furthermore, we demonstrate that SNHG12 could promote the progression of DLBCL cells by targeting miR-195. Our findings suggest that SNHG12 is involved in DLBCL tumorigenesis and is a novel biomarker that can be used as a therapeutic candidate for DLBCL patients.
Patients and Methods
Patients and Clinical Samples
After obtention of patients’ written informed consent, a total of 80 biopsy tumor samples were collected form patients with activated B-cell like DLBCL at the 2nd Affiliated Hospital of Harbin Medical University (Harbin, China) from 2015 to 2018. The other 80 control biopsy specimens were obtained from patients with reactive lymphoid hyperplasia (RLH), who were suspected of lymphoma during the same period. The specimens were stored at −80°C until use. None of the patients underwent previous diagnostic biopsies, and all them were confirmed by two independent pathologists. Some clinical parameters, including age, gender, B symptoms, clinical stage, extra-nodal invasion, serum lactate dehydrogenase (LDH) level and International Prognostic Index (IPI) score were recorded. All DLBCL patients were enrolled into the follow up survival survey, which was censored on September 2019. The Overall survival (OS) was calculated from the time of resection to the time of death or last follow-up. Disease-free survival (DFS) was calculated from the time of resection to the first time of recurrence diagnosis, death or last follow-up. The protocol for tissue collection was approved by the Ethics Committee of the 2nd Affiliated Hospital of Harbin Medical University (Harbin, China). The study was conducted in compliance with the Declaration of Helsinki and the guidelines of the committee.
Cell Culture
The human DLBCL cell lines OCI-LY7 and OCI-LY3, and the normal B lymphocytes IM-9I were purchased from the Cell Bank of Type Culture Collection of the Chinese Academy of Sciences (Shanghai, China). OCI-LY7 and OCI-LY3 cell lines were grown in IMEM medium (Gibco) that contained 10% fetal bovine serum (FBS; Invitrogen). IM-9I cells were cultured in RPMI 1640 culture medium (Gibco) supplemented with 10% FBS (Invitrogen). Cells were maintained at 37°C and in a humidified atmosphere containing 5% CO2.
RNA Preparation and Quantitative Real-Time PCR (qRT-PCR) Detection
Total RNA was extracted using TRIzol reagent (Invitrogen; Thermo Fisher Scientific, Inc.) and reverse-transcribed to cDNA using the PrimeScript RT reagent kit (Takara Bio, Inc.), according to the manufacturer’s instructions. SYBR® Premix Ex Taq (Takara Bio, Inc.) was used for qRT-PCR. The thermocycling conditions used for the PCR were as follows: 95°C for 1 min; 40 cycles of 95°C for 12 sec and 58.5°C for 40 sec. SNHG12 and miR-195 Fold changes were normalized to GAPDH and U6, respectively and using the 2−ΔΔCt method. All used PCR primers are shown in Table 1.
 |
Table 1 The Sequences of PCR Primers |
Cell Transfection
SNHG12 small interfering RNA (siRNA) and the non-targeting control sequence (si-NC) were synthesized by Sangon Biotech Co., Ltd. The SNHG12 expressing plasmid was constructed by inserting the full-length human SNHG12 sequence into pcDNA3.1 vector from Sangon Biotech Co., Ltd. MiR-195 mimics (#HMI0320) and the scramble miRNA control (miR-NC) (#HMI0321) were purchased from Sigma-Aldrich (Sigma, USA). Cell transfections were performed using Lipofectamine 2000 reagent (Invitrogen) and at appropriate concentrations.
Cell Proliferation Assay
Cell proliferation was measured using the cell counting kit-8 (CCK-8, #C0037, Beyotime, Shanghai, China). A total of 5 × 103 cells was seeded in each well of a 96-well plate, and incubated for 24 h. Ten Microliter WST-8 from the CCK-8 Kit was added to each well and the optical density was measured at 450 nm using a microtiter plate reader at 0, 1, 2, 3 and 4 days.
Transwell Assay
A 24-well Transwell assay (#3395, 8.0μm, Corning) was used to examine the migration and invasion abilities of DLBCL cells. For the migration assay, 150μL cell suspension (1 × 105 cells) was seeded into the upper chamber. For the invasion assay, the upper chamber was precoated with Matrigel (BD Biosciences). Next, 500 μL IMEM medium with 10% FBS was placed into the lower chamber. The cells were maintained at 37°C in a humidified atmosphere with 5% CO2. After 48 hours incubation, invasive cells were fixed with 100% methanol at 4°C for 15 min and stained with 0.1% crystal violet at 4°C for 10min. Five fields were randomly selected and the cells were counted under a light microscope (Olympus Corporation) at ×200 magnification.
Tumorigenesis in vivo
A total of 8 male BALB/c nude mice (~5 weeks old) were raised under standard conditions. OCI-LY7 cells (1×106 cells/mouse), transfected with si-SNHG12 or si-NC, were injected into the right flanks of the nude mice (4 mice per group). The mice were monitored, and tumors’ volume weekly measured using the simple formula (0.5 × length × width2). After 4 weeks, the mice were sacrificed, the tumors excised, photographed and weighed. The in vivo xenograft assay was approved by the Animal Ethical and Welfare Committee of the 2nd Affiliated Hospital of Harbin Medical University (Harbin, China). The study was performed in accordance with NC3Rs ARRIVE guidelines.18
RNA Immunoprecipitation (RIP) Assay
The RIP assay was conducted using the Magna RIP Kit (#17-700, EMD Millipore). Human anti-Ago2 antibodies (#06-642, Millipore) were used to capture the RNAs used for qRT-PCR analysis. SNHG12 or miR-195 expression were measured by qRT-PCR.
Luciferase Reporter Assay
Cells were placed in a 24-well plate and co-transfected with SNHG12 mutant (MUT) or wild-type (WT) SNHG12, and miRNAs (miR-195 mimic or mi-NC) using Lipofectamine® 2000 (Invitrogen; Thermo Fisher Scientific, Inc.). After 24 h of incubation, the cells were lysed, and reporter activities were determined using a dual-luciferase reporter assay system (Promega Corporation).
Statistical Analysis
All data were analyzed with GraphPad Prism 5 (GraphPad Software Inc., CA, USA) and the SPSS 20 software package (IBM SPSS Inc, Chicago, IL). All results were expressed as mean ± standard deviation. Student’s t-test or one-way ANOVA analysis were used to analyze the difference. Categorical data were compared using the chi-square test and Pearson’s correlation analysis was applied to analyze the correlations between SNHG12 and miR-195. Kaplan–Meier curves were generated for overall survival (OS) and disease-free survival (DFS). A p-value of < 0.05 was considered as statistically significant.
Results
Clinical Significance of SNHG12 Expression in DLBCL
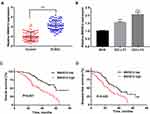
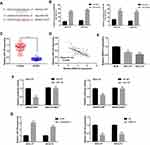
SNHG12 expression was first determined by qRT-PCR analysis of tissue samples from 80 DLBCL and RLH patients, and we found that SNHG12 was considerably increased in DLBCL tissues compared with RLH tissues (p < 0.001, Figure 1A). Next, we examined SNHG12 expression in the human DLBCL cell lines OCI-LY7 and OCI-LY3 and in normal B lymphocytes IM-9I, and the results revealed that SNHG12 was overexpressed in the DLBCL cells compared with IM-9I cells (p < 0.001, Figure 1B). Furthermore, using SNHG12 expression median value as cut-off value, we classified DLBCL patients into two groups: SNHG12 low (below the median, 40 patients) and SNHG12 high (above the median, 40 patients). The results showed that DLBCL patients, with high SNHG12 expression, had worse OS and DFS that those with lower SNHG12 expression (Figure 1C and D; p = 0.001 and p = 0.023, respectively). Collectively, all the above results showed that SNHG12 was highly expressed in DLBCL tissues and this correlated with patients’ poor prognosis.
In this group of 80 DLBCL patients, the relationship between SNHG12 expression and clinicopathologic parameters, was also explored. As shown in Table 2, the number of patients with high SNHG12 expression was higher in clinical stages ш-Ⅳ (p = 0.003). In the high SNHG12 expression group the proportion of extra-nodal invasion was higher compared with the low SNHG12 expression group (p = 0.012). The high SNHG12 expression group had more patients with a serum LDH ≥ 300, while the low SNHG12 expression group had more patients with LDH < 300 (p = 0.014). In addition, the multivariate analysis showed that SNHG12 expression was a significant prognostic factor both for OS (p = 0.003, Table 3) and DFS (p = 0.021, Table 4).
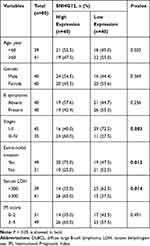 |
Table 2 SNHG12 Expression and Clinicopathologic Features in 80 Cases of DLBCL |
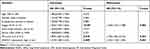 |
Table 3 Univariate and Multivariate Analyses for Overall Survival in 80 Cases of DLBCL |
 |
Table 4 Univariate and Multivariate Analyses for Disease-Free Survival in 80 Cases of DLBCL |
Downregulation of SNHG12 Inhibits the Growth, Migration and Invasion of DLBCL Cells in vitro
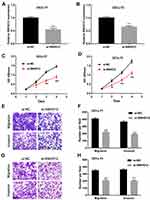
Based on the above clinical findings, experiments were further conducted to investigate the biological function of SNHG12 in DLBCL growth and metastasis in vitro. To downregulate SNHG12 expression, siRNA and non-targeting control sequence (si-NC) were transfected into OCI-LY7 and OCI-LY3 cells. The efficiency of SNHG12 knockdown was confirmed by qRT-PCR (p < 0.001, Figure 2A and B). Subsequently, the effects of SNHG12 on the proliferation of DLBCL cells were assessed by the CCK-8 assay, and the results showed that SNHG12 knockdown significantly inhibited the growth abilities of both OCI-LY7 and OCI-LY3 cells (p < 0.05, Figure 2C and D). In addition, the migration and invasion abilities of OCI-LY7 and OCI-LY3 cells, assessed by the transwell assay, were significantly inhibited following SNHG12 downregulation (p < 0.05, Figure 2E–H). Taken together, these results demonstrate that SNHG12 downregulation inhibits the growth, migration, and invasion abilities of DLBCL cells in vitro.
SNHG12 Overexpression Promotes the Growth, Migration and Invasion of DLBCL Cells in vitro
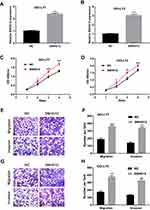
To further evaluate the function of SNHG12 in DLBCL cells, pcDNA3.1-SNHG12 was transfected in OCI-LY7 and OCI-LY3 cells and the efficiency of transfection was confirmed by qRT-PCR (Figure 3A and B). The results of CCK-8 assay demonstrated that SNHG12 overexpression significantly promoted the growth abilities of OCI-LY7 and OCI-LY3 cells (p < 0.05, Figure 3C and D). In addition, the transwell assay indicated that SNHG12 overexpression promotes the migration and invasion of OCI-LY7 and OCI-LY3 cells (Figure 3E–H). These results demonstrate that the SNHG12 overexpression promotes the growth, migration, and invasion of DLBCL cells in vitro.
SNHG12 Knockdown Suppresses Tumorigenesis of DLBCL Cells in vivo
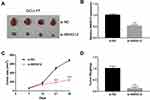
To investigate SNHG12 role in the growth of DLBCL cells in vivo, xenograft mouse models were established using OCI-LY7 control and SNHG12 knockdown cells. The results showed that SNHG12 downregulation inhibits tumor growth after 4 weeks (Figure 4A). QRT-PCR analysis was performed to confirm SNHG12 downregulation in the SNHG12 knockdown tumors compared to control tumors (Figure 4B). Moreover, the tumors in the SNHG12 knockdown group grew slower and were lighter compared to the control group tumors (n = 4, p < 0.001) (Figure 4C and D).
SNHG12 Serves as a Sponge for miR-196 in DLBCL
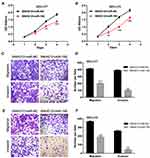
As predicted by the online software Starbase v2.0 (http://starbase.sysu.edu.cn/starbase2/), SNHG12 contains one miR-195 conserved target binding site (Figure 5A). SNHG12 localization has been reported to be mainly cytoplasmic;17 thus, we supposed that SNHG12 may form an RNA-induced silencing complex (RISC) with miRNAs in DLBCL. To investigate this possibility, a RIP assay was conducted using human anti-Ago2 antibodies and that showed a significant enrichment of SNHG12 in DLBCL cells (Figure 5B). Using the same assay, we also found a significant enrichment of miR-129-5p in DLBCL cells (Figure 5B). The expression of miR-195 showed a decreasing trend in DLBCL tissues compared with RLH tissues (p < 0.001, Figure 5C). Pearson correlation analysis demonstrated that SNHG12 expression negatively correlates with miR-195 expression in DLBCL tissues (p < 0.001, Figure 5D). Besides, miR-195 had lower expression levels in DLBCL cells compared to IM-9I (p < 0.001, Figure 5E). To investigate potential physical interactions between SNHG12 and miR-195, a luciferase reporter assay was conducted in OCI-LY9 and OCI-LY3 cells that were co-transfected with the WT or MUT SNHG12 reporters and miR-195 mimic or mi-NC. The results showed that miR-195 overexpression results in a significant reduction of the WT reporter activity in OCI-LY9 and OCI-LY3 cells. Yet, the mutant reporter activities in all transfected cells showed no significant changes (Figure 5F). These results indicated that SNHG12 and miR-195 interaction was an independently regulatory factor. In addition, si-SNHG12 transfected cells showed an increased expression of miR-195, while the cells transfected with pcDNA-SNHG12 had a decreased expression of miR-195 (Figure 5G). Collectively, these results showed that SNHG12 directly suppresses the expression of miR-195 by sponging miR-195 in DLBCL.
To determine whether miR-195 is involved in SNHG12 oncogenic role in DLBCL, we transfected miR-195 mimic or mi-NC in SNHG12 overexpressing OCI-LY9 and OCI-LY3 cells. As demonstrated in Figure 6A–F, miR-195 upregulation rescued the effects of SNHG12 on the proliferation, migration and invasion of DLBCL cells. This result indicates that miR-195 prevents the proliferation, migration and invasion capabilities of DLBCL cells by interacting with SNHG12.
Discussion
Several studies have demonstrated that SNHG12 plays a significant oncogenic role in various cancers. Specifically, SNHG12 was shown to act as an oncogene by regulating the expression of Notch2 through miR-195-5p sponging in osteosarcoma.19 SNHG12 was also reported to promote multidrug resistance via the SNHG12-miR-181a-MAPK/Slug axis.20 Additionally, SNHG12 can also promote the growth and metastasis of papillary thyroid carcinoma cells via regulating the Wnt/β-catenin signaling pathway.21 However, the potential biological function of SNHG12 in DLBCL progression is unknown.
In this study, we revealed that SNHG12 expression is upregulated in DLBCL tissues compared with that in RLH tissues. DLBCL patients with high SNHG12 expression, exhibited poor OS and DFS compared to those with low SNHG12 expression, indicating that SNHG12 could be a DLBCL prognostic factor. Moreover, we found that SNHG12 expression levels correlate with patients’ clinical characteristics, such as clinical stage, extra-nodal invasion and LDH serum level. All the above clinical results indicated that SNHG12 may play a critical role in DLBCL progression. Next, SNHG12 biological functions in DLBCL were evaluated, and the results demonstrated that SNHG12 knockdown suppresses the proliferative, migratory and invasive capacities of DLBCL cells in vitro. In contrast, SNHG12 upregulation promoted DLBCL cell proliferation, migration, and invasion. Furthermore, SNHG12 knockdown restrained tumorigenesis of DLBCL cells in the xenograft mouse models.
Subsequently, we identified a target binding site of miR-195 in SNHG12 using bioinformatics analysis, that prompted investigations on a potential ceRNA mechanism that involved SNHG12 and miR-195. To this end, we first confirmed that miR-195 could bind SNHG12 by conducting a dual luciferase assay. Next, the negative correlation between SNHG12 and miR-195 was confirmed in DLBCL cells and clinical tissues. We also showed that miR-195 upregulation abolishes the effects of SNHG12 on DLBCL cell proliferation, migration and invasion. MiR-195 is originated in intron 7 and located on human chromosome 17p13.1, which was reported to be involved in various diseases, including heart failure, schizophrenia, and cancer.22 MiR-195 has been reported to be involved in the progression of cancer by regulating some key genes such as CDK6, Bcl-2 and WEE1. The dysregulation and tumor suppressor role of miR-195 has been determined in various cancers. For example, one meta-analysis23 revealed that elevated level of miRNA-195 expression serves as a potential biomarker that predicts favorable prognoses for various cancers in China, including non-small cell lung cancer,24 gastric cancer,25 hepatocellular cancer26 and colorectal cancer.27 Moreover, miR-195 could suppress the progression of prostate cancer, lung adenocarcinoma and endometrial carcinoma.28–30 A study reported that LINC00355 can sponge miR-195, which inhibits apoptosis promotes the progression of head and neck squamous cell carcinoma.31 Wu et al demonstrated that BRAF-activated noncoding RNA promotes tumorigenesis of pancreatic cancer by sponging miR-195.32 Overall, our results demonstrated that SNHG12 could accelerate tumorigenesis of DLBCL cells through miR-195 sponging.
To conclude, this study provides new insights into the role of SNHG12 in DLBCL progression through a mechanism involving miR-195 sponging. SNHG12 expression predicts poor clinical outcome and serves as a novel oncogene in DLBCL. These findings also suggest that SNHG12 may serve as a DLBCL potential therapeutic target.
Disclosure
The authors report no conflicts of interest in this work.
References
1. Luminari S. Bridging the gap between epidemiology and clinical research in lymphoma. Leuk Lymphoma. 2013;54(9):1855–1856. doi:10.3109/10428194.2013.777838
2. Smedby KE, Hjalgrim H. Epidemiology and etiology of mantle cell lymphoma and other non-Hodgkin lymphoma subtypes. Semin Cancer Biol. 2011;21(5):293–298. doi:10.1016/j.semcancer.2011.09.010
3. Martelli M, Ferreri AJ, Agostinelli C, Di Rocco A, Pfreundschuh M, Pileri SA. Diffuse large B-cell lymphoma. Crit Rev Oncol Hematol. 2013;87(2):146–171. doi:10.1016/j.critrevonc.2012.12.009
4. Jiang W, Huang H. The evolution and curative effect of diffuse large B cell lymphoma treatment in China. Zhonghua Xue Ye Xue Za Zhi. 2014;35(4):357–360. doi:10.3760/cma.j.issn.0253-2727.2014.04.025
5. Castillo JJ, Winer ES, Olszewski AJ. Sites of extranodal involvement are prognostic in patients with diffuse large B-cell lymphoma in the rituximab era: an analysis of the surveillance, epidemiology and end results database. Am J Hematol. 2014;89(3):310–314. doi:10.1002/ajh.23638
6. Saito B, Shiozawa E, Usui T, et al. Rituximab with chemotherapy improves survival of non-germinal center type untreated diffuse large B-cell lymphoma. Leukemia. 2007;21(12):2563–2566. doi:10.1038/sj.leu.2404844
7. Gisselbrecht C. Use of rituximab in diffuse large B-cell lymphoma in the salvage setting. Br J Haematol. 2008;143(5):607–621. doi:10.1111/j.1365-2141.2008.07383.x
8. Jarroux J, Morillon A, Pinskaya M. History, discovery, and classification of lncRNAs. Adv Exp Med Biol. 2017;1008:1–46.
9. Mathy NW, Chen XM. Long non-coding RNAs (lncRNAs) and their transcriptional control of inflammatory responses. J Biol Chem. 2017;292(30):12375–12382. doi:10.1074/jbc.R116.760884
10. Bhan A, Soleimani M, Mandal SS. Long noncoding RNA and cancer: a new paradigm. Cancer Res. 2017;77(15):3965–3981. doi:10.1158/0008-5472.CAN-16-2634
11. Qu L, Ding J, Chen C, et al. Exosome-transmitted lncARSR promotes sunitinib resistance in renal cancer by acting as a competing endogenous RNA. Cancer Cell. 2016;29(5):653–668. doi:10.1016/j.ccell.2016.03.004
12. Wu Z, Chen D, Wang K, Cao C, Xu X. Long non-coding RNA SNHG12 functions as a competing endogenous RNA to regulate MDM4 expression by sponging miR-129-5p in clear cell renal cell carcinoma. Front Oncol. 2019;9:1260. doi:10.3389/fonc.2019.01260
13. Cheng G, Song Z, Liu Y, et al. Long noncoding RNA SNHG12 indicates the prognosis of prostate cancer and accelerates tumorigenesis via sponging miR-133b. J Cell Physiol. 2020;235(2):1235–1246. doi:10.1002/jcp.29039
14. Jin XJ, Chen XJ, Zhang ZF, et al. Long noncoding RNA SNHG12 promotes the progression of cervical cancer via modulating miR-125b/STAT3 axis. J Cell Physiol. 2019;234(5):6624–6632. doi:10.1002/jcp.27403
15. Liu ZB, Tang C, Jin X, Liu SH, Pi W. Increased expression of lncRNA SNHG12 predicts a poor prognosis of nasopharyngeal carcinoma and regulates cell proliferation and metastasis by modulating Notch signal pathway. Cancer Biomarkers. 2018;23(4):603–613. doi:10.3233/CBM-181873
16. Wang JZ, Xu CL, Wu H, Shen SJ. LncRNA SNHG12 promotes cell growth and inhibits cell apoptosis in colorectal cancer cells. Braz J Med Biol Res. 2017;50(3):e6079. doi:10.1590/1414-431X20176071
17. Lan T, Ma W, Hong Z, Wu L, Chen X, Yuan Y. Long non-coding RNA small nucleolar RNA host gene 12 (SNHG12) promotes tumorigenesis and metastasis by targeting miR-199a/b-5p in hepatocellular carcinoma. J Exp Clin Cancer Res. 2017;36(1):11. doi:10.1186/s13046-016-0486-9
18. Kilkenny C, Browne WJ, Cuthill IC, Emerson M, Altman DG. Improving bioscience research reporting: the ARRIVE guidelines for reporting animal research. PLoS Biol. 2010;8(6):e1000412. doi:10.1371/journal.pbio.1000412
19. Zhou S, Yu L, Xiong M, Dai G. LncRNA SNHG12 promotes tumorigenesis and metastasis in osteosarcoma by upregulating Notch2 by sponging miR-195-5p. Biochem Biophys Res Commun. 2018;495(2):1822–1832. doi:10.1016/j.bbrc.2017.12.047
20. Wang P, Chen D, Ma H, Li Y. LncRNA SNHG12 contributes to multidrug resistance through activating the MAPK/Slug pathway by sponging miR-181a in non-small cell lung cancer. Oncotarget. 2017;8(48):84086–84101. doi:10.18632/oncotarget.20475
21. Ding S, Qu W, Jiao Y, Zhang J, Zhang C, Dang S. LncRNA SNHG12 promotes the proliferation and metastasis of papillary thyroid carcinoma cells through regulating wnt/β-catenin signaling pathway. Cancer Biomarkers. 2018;22(2):217–226. doi:10.3233/CBM-170777
22. Flavin RJ, Smyth PC, Laios A, et al. Potentially important microRNA cluster on chromosome 17p13.1 in primary peritoneal carcinoma. Mod Pathol. 2009;22(2):197–205. doi:10.1038/modpathol.2008.135
23. Wang N, Cao Y, Ge X, et al. MicroRNA-195 as a prognostic factor for cancer survival outcome in China: a meta-analysis. Cancer Manag Res. 2019;11:7967–7979. doi:10.2147/CMAR.S205841
24. Liu B, Qu J, Xu F, et al. MiR-195 suppresses non-small cell lung cancer by targeting CHEK1. Oncotarget. 2015;6(11):9445–9456. doi:10.18632/oncotarget.3255
25. Shen YH, Xie ZB, Yue AM, et al. Expression level of microRNA-195 in the serum of patients with gastric cancer and its relationship with the clinicopathological staging of the cancer. Eur Rev Med Pharmacol Sci. 2016;20(7):1283–1287.
26. Yu S, Jing L, Yin XR, et al. MiR-195 suppresses the metastasis and epithelial-mesenchymal transition of hepatocellular carcinoma by inhibiting YAP. Oncotarget. 2017;8(59):99757–99771. doi:10.18632/oncotarget.20909
27. Wang X, Wang J, Ma H, Zhang J, Zhou X. Downregulation of miR-195 correlates with lymph node metastasis and poor prognosis in colorectal cancer. Med Oncol. 2012;29(2):919–927. doi:10.1007/s12032-011-9880-5
28. Liu C, Guan H, Wang Y, et al. miR-195 inhibits EMT by targeting FGF2 in prostate cancer cells. PLoS One. 2015;10(12):e0144073. doi:10.1371/journal.pone.0144073
29. Zhou Y, Zhao M, Du Y, et al. MicroRNA-195 suppresses the progression of lung adenocarcinoma by directly targeting apelin. Thorac Cancer. 2019;10(6):1419–1430. doi:10.1111/1759-7714.13087
30. Deng J, Wang W, Yu G, Ma X. MicroRNA195 inhibits epithelial-mesenchymal transition by targeting G protein-coupled estrogen receptor 1 in endometrial carcinoma. Mol Med Rep. 2019;20(5):4023–4032. doi:10.3892/mmr.2019.10652
31. Lu S, Sun Z, Tang L, Chen L. LINC00355 promotes tumor progression in HNSCC by hindering microRNA-195-mediated suppression of HOXA10 expression. Mol Ther Nucleic Acids. 2019;19:61–71. doi:10.1016/j.omtn.2019.11.002
32. Wu X, Xia T, Cao M, et al. LncRNA BANCR promotes pancreatic cancer tumorigenesis via modulating MiR-195-5p/Wnt/β-catenin signaling pathway. Technol Cancer Res Treat. 2019;18:1533033819887962. doi:10.1177/1533033819887962
 © 2020 The Author(s). This work is published and licensed by Dove Medical Press Limited. The full terms of this license are available at https://www.dovepress.com/terms.php and incorporate the Creative Commons Attribution - Non Commercial (unported, v3.0) License.
By accessing the work you hereby accept the Terms. Non-commercial uses of the work are permitted without any further permission from Dove Medical Press Limited, provided the work is properly attributed. For permission for commercial use of this work, please see paragraphs 4.2 and 5 of our Terms.
© 2020 The Author(s). This work is published and licensed by Dove Medical Press Limited. The full terms of this license are available at https://www.dovepress.com/terms.php and incorporate the Creative Commons Attribution - Non Commercial (unported, v3.0) License.
By accessing the work you hereby accept the Terms. Non-commercial uses of the work are permitted without any further permission from Dove Medical Press Limited, provided the work is properly attributed. For permission for commercial use of this work, please see paragraphs 4.2 and 5 of our Terms.