Back to Journals » OncoTargets and Therapy » Volume 13
Long-Noncoding RNA PCAT6 Aggravates Osteosarcoma Tumourigenesis via the MiR-143-3p/ZEB1 Axis
Authors Wu K, Feng Q, Li L, Xiong Y, Liu S, Liu J, Wu Q
Received 22 April 2020
Accepted for publication 8 August 2020
Published 26 August 2020 Volume 2020:13 Pages 8705—8714
DOI https://doi.org/10.2147/OTT.S258415
Checked for plagiarism Yes
Review by Single anonymous peer review
Peer reviewer comments 2
Editor who approved publication: Prof. Dr. Nicola Silvestris
Kai Wu,1,* Qiong Feng,2,* Liang Li,1 Yanfei Xiong,3 Shihong Liu,4 Jie Liu,1 Qing Wu1
1Department of Orthopedics, The Second Affiliated Hospital of Nanchang University, Nanchang 300006, People’s Republic of China; 2Nursing School, Nanchang University, Nanchang 300006, People’s Republic of China; 3Department of Orthopedics, Jingan Hospital, Yichun 330600, People’s Republic of China; 4Jingan Hospital of Traditional Chinese Medicine, Yichun 330600, People’s Republic of China
*These authors contributed equally to this work
Correspondence: Qing Wu Email [email protected]
Introduction: The long-noncoding RNA PCAT6 plays an important regulatory role in the development of several cancers. However, the expression pattern and underlying mechanisms of PCAT6 in osteosarcoma (OS) are yet unknown.
Methods: We used real-time PCR to measure PCAT6 expression in 106 tumor pairs and corresponding non-tumor tissues from OS patients. Statistical analyses were applied to evaluate the prognostic value and associations of PCAT6 expression with clinical parameters. Furthermore, the PCAT6 was silenced with siRNA in OS cells. Moreover, phenotype of PCAT6 silenced OS cells was measured using colony formation, CCK-8, cell migration and invasion assay. Finally, the molecular mechanism of PCAT6/miR-143-3p/ZEB1 axis in OS progression was explored.
Results: The expression level of PCAT6 in OS tissues was significantly elevated as compared with that in the adjacent normal bone tissues and that high PCAT6 expression closely correlated with the malignant phenotype and poor survival among patients with OS. Multivariate analyses revealed PCAT6 overexpression as an independent prognostic factor for the poor outcome of patients with OS. Functional assay results demonstrated that the knockdown of PCAT6 expression notably suppressed the proliferation, migration, and invasion of OS cells. An elevated PCAT6 level aggravated the malignant phenotype of OS cells via ZEB1 expression upregulation. Mechanistic studies revealed that PCAT6 could sponge endogenous miR-143-3p and inhibit its activity, resulting in an increase in ZEB1 level. Finally, we demonstrated that the tumour-promoting role of PCAT6 in OS was dependent on the regulation of the miR-143-3p/ZEB1 axis.
Conclusion: These findings highlight the potential role of PCAT6, which could serve as a valuable prognostic indicator for patients with OS.
Keywords: osteosarcoma, PCAT6, ZEB1, miR-143-3p
Introduction
Osteosarcoma (OS) is the most common type of malignant tumour of the skeletal system.1,2 Despite the wide use of surgery and systemic chemotherapy, the overall 5-year survival rate remains very poor owing to metastasis.3,4 Thus, exploration of the molecular mechanism underlying the carcinogenesis and progression of OS is desirable.
Long-noncoding RNAs (lncRNAs) are more than 200 nucleotides in length but lack any evident protein-coding functions.5 LncRNAs are emerging as crucial regulators of various biological processes6 and are involved in the development of diseases, especially cancer.7,8 Evidence indicates that lncRNAs are commonly dysregulated in a variety of cancers9,10 and play crucial roles in tumourigenesis.11 The lncRNA prostate cancer-associated transcript 6 (PCAT6) was reported to enhance the growth of prostate cancer cells in an androgen-independent manner.12 Recent studies have revealed the more ubiquitous expression of PCAT6 and its influence on the progression of several cancers.13–15 For instance, PCAT6 is overexpressed in gastric cancer tissues and contributes to gastric cancer progression by endogenously competing with microRNA (miR)-30.15 However, the clinical significance and potential role of PCAT6 in OS are still unknown.
In the present study, we found that high PCAT6 level was closely related to the progression of OS and poor patient outcome. Our findings also indicate that PCAT6 expression knockdown suppressed the growth and metastasis of OS cells and that PCAT6 exerted its function by competitively sponging and subsequently inhibiting miR-143-3p, a crucial repressor of tumour development in different cancers,16–18 resulting in an increase in ZEB1 level. Thus, our study identifies PCAT6 as a novel regulator of OS development and provides a promising prognostic and therapeutic target for OS.
Materials and Methods
Patients and Tumour Specimens
We obtained 106 pairs of primary OS tumour tissues and corresponding normal bone tissues from patients undergoing resection surgery at the Department of Orthopedics, the Second Affiliated Hospital of Nanchang University from 2013 to 2018. All specimens from resection surgery were frozen and stored at −80°C for further analysis. The clinical characteristics of all patients are summarised in Table 1. Written informed consent was obtained from each patient, and the study was approved by the Ethic and Research Committees of the Second Affiliated Hospital of Nanchang University.
 |
Table 1 Relationship Between PCAT6 Expression and Clinicopathological Features |
Cell Lines and Culture Conditions
Four human OS cell lines (MG-63, Saos-2, 143B, and U2OS) and the normal human osteoblast line hFOB 1.19 were purchased from the American Type Culture Collection (Manassas, USA). The cells were cultured in Dulbecco’s modified Eagle’s medium (DMEM; Gibco) supplemented with foetal bovine serum (FBS; HyClone) at a final concentration of 10% at 37°C under 5% CO2 in a humidified chamber.
RNA Extraction and Real-Time Quantitative Polymerase Chain Reaction (RT-qPCR)
The RT-qPCR assay was performed according to a previously described protocol with minor revisions.19 The primers were designed as follows: PCAT6 forward primer, 5-CCCCTCCTTACTCTTGGACAAC-3 and reverse primer, 5- GACCGAATGAGGATGGAGACAC-3; ZEB1 forward primer, 5-AGCGAGGTAAAGTTGCGTCT-3 and reverse primer, 5-AGGTTTTCTGGGCCATACCG-3; GAPDH forward primer, 5-ATGGGCAGCCGTTAGGAAAG-3 and reverse primer 5-ATCACCCGGAGGAGAAATCG-3. Hsa-miR-143-3p primers were purchased from RiboBio. RNU6B was used as an internal control for RT-qPCR.
Western Blot Analysis
Western blot analysis to detect specific protein expression was carried out using the following antibodies: anti-ZEB1 (1:1000, Abcam) and anti-Tubulin (1:1000, Abcam). The signals were detected with enhanced chemiluminescence.
PCAT6 Small-Interfering RNA (siRNA), Short-Hairpin RNA (shRNA) and Plasmids
The siRNAs or the RNA duplexes specifically targeting PCAT6 were synthesised by GenePharma (Shanghai, China). The nonspecific siRNA oligonucleotides were used as negative controls. In order to obtain stable PCAT6 knockdown cell lines, the oligoduplexes were cloned into pLKO.1-TRC cloning vector and transfected into OS cells. Then, the transfected cells were selected by incubation with 2μg/mL of puromycin for 2 weeks. Additionally, the Full-length human ZEB1 cDNA was compounded by Genepharma and ligated into pcDNA3.1 vector (p-ZEB1). An empty vector was used as the negative control.
Cell Proliferation and Cell Cycle Assays
To examine cell proliferation, 5x103 cells/well were seeded in a 96-well plate. At the indicated time points, viable cells were counted using the Cell Counting Kit-8 (CCK-8; Dojindo Laboratories, USA) or colony formation assay. In addition, cell cycle assays were performed as previously described.19
Cell Migration and Invasion Assay
The migration ability of OS cells was routinely tested in a Transwell Boyden Chamber (8-mm pore size, BD Biosciences). For the cell invasion assay, the polycarbonate membranes of the upper compartment of the chambers were precoated with a matrix gel.
Nude Mouse Model of Tibia Orthotopic Tumour
The in vivo xenograft studies were carried out as previously described.20 All animal experiments were approved by the Animal Care and Use Committee of Nanchang University and performed in accordance with the guidelines of the Animal Care and Use Committee of Nanchang University (NIH publication 85–23, revised 1985). Lung tissues were harvested, fixed in 10% formalin, and embedded in paraffin. Metastases in the lung were examined by gross observation and histological assessment.
Protein Identification and Liquid Chromatography–Tandem Mass Spectrometry (LC-MS/MS) Analysis
Isobaric tags for relative and absolute quantitation (iTRAQ) and LC-MS/MS analysis were performed as previously described with minor modifications.21
Dual-Luciferase Reporter and RNA Immunoprecipitation (RIP) Assays
For reporter assays, PCAT6 3ʹ-UTR-wt and PCAT6 3ʹ-UTR-mut were cloned into the PmirGLO Luciferase plasmids GenePharma (shanghai, China), which were transfected into OS cells using Lipofectamine 2000. After incubation for 48 h, the luciferase activity was measured with Dual-Luciferase Reporter Assay System (Promega, China) according to the manufacturer’s instruction. RIP assays were performed as previously described.20
Statistical Analysis
All results are shown as mean±standard deviation (SD) and analysed using GraphPad Prism 6 (GraphPad Software, USA) from at least three independent experiments. The Kaplan–Meier method was used to generate the survival curve, and the Log rank test was used to determine statistical significance. The differences between groups were analysed with two-tailed Student’s t-test and analysis of variance (ANOVA). Data were considered statistically significant at P< 0.05.
Results
PCAT6 Expression is Upregulated in Human OS Tissues and Closely Correlates with Clinical Outcomes
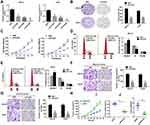
To investigate the expression of PCAT6 in OS, 106 cases of primary OS and corresponding normal bone tissues were analysed. RT-qPCR data showed that the expression level of PCAT6 in tumour tissues was significantly higher than that in normal bone tissues (Figure 1A and B). The expression of PCAT6 in OS cells was always higher than that in normal human osteoblasts (Figure 1C).
The correlation between PCAT6 expression and clinical parameters was evaluated. As shown in Table 1, the overexpression of PCAT6 significantly correlated with the size of large tumours (P=0.009), Enneking stage (P<0.0001), and distant metastasis (P<0.0001). In addition, multivariate Cox regression analysis revealed PCAT6 overexpression as an independent predictor of low survival in patients with OS (Table 2). Furthermore, Kaplan–Meier survival curve showed that patients with high expression of PCAT6 had worse overall and progression-free survival than those with low PCAT6 expression (Figure 1D and E). Overall, these results suggest that the overexpression of PCAT6 in OS tissues may serve as a valuable prognostic marker for OS.
 |
Table 2 Univariate and Multivariate Analyses of Overall Survival in OS Patients (Cox Proportional Hazards Regression Model) |
PCAT6 Silencing Suppresses the Proliferation, Migration, and Invasion of OS Cells
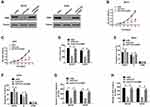
To study the biological function of PCAT6 in OS, we silenced its expression in MG-63 and U2OS cells (Figure 2A). As shown in Figure 2B, the result of colony formation analysis showed that PCAT6 knockout reduced the clonal survival rates of MG-63 and U2OS cells. CCK-8 assay results also showed that the silencing of PCAT6 expression significantly inhibited the proliferation of MG-63 and U2OS cells (Figure 2C). In comparison with the control group, the group subjected to PCAT6 silencing showed a significant increase in the number of cells at the G0/G1 phase and a decrease in the cells in S phase (Figure 2D and E).
We performed cell migration and invasion experiments to explore the effect of PCAT6 on tumour metastasis. As shown in Figure 2F and G, the knockout of PCAT6 expression significantly inhibited the invasion and migration of MG-63 and U2OS cells. In vivo experiments showed that PCAT6 expression silencing significantly inhibited the growth of OS cells as compared with the control cells (Figure 2H and I). PCAT6 knockout also significantly reduced the incidence and number of lung metastases as compared with the control group (Figure 2J). These data suggest that PCAT6 knockout inhibits the tumourigenicity of OS cells in vitro and in vivo. Overall, our results show that PCAT6 may play the role of a key carcinogenic factor in OS tumourigenesis.
PCAT6 Promotes the Progression of OS Through Upregulating ZEB1 Expression
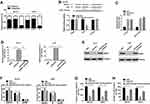
To explore the molecular mechanism underlying the effect of PCAT6 on promoting OS progression, we used iTRAQ and MS to identify the proteins expressed at different levels in MG-63 cells subjected to PCAT6 expression silencing. MS analysis revealed a significant change in ZEB1 level in cells (Figure 3A). Consistently, Western blot results confirmed that the deletion of PCAT6 in MG-63 and U2OS cells resulted in the reduction in ZEB1 protein levels as compared with that in the control cells (Figure 3B). We also investigated whether PCAT6 upregulates the expression of ZEB1 in a transcription-dependent manner. As expected, the mRNA level of ZEB1 reduced in OS cells treated with shPCAT6 (Figure 3C and D). Additionally, the level of ZEB1 protein in OS tissues was significantly higher than that in normal bone tissues (Figure 3E), and the protein level of ZEB1 positively correlated with the expression of PCAT6 in OS tissues (Figure 3F). Thus, these findings suggest that PCAT6 promotes the progression of OS through upregulating ZEB1 expression.
The Oncogenic Function of PCAT6 in OS Cells in vitro Was Dependent on ZEB1
We performed rescue experiments to determine whether PCAT6 knockdown influenced OS cell proliferation, cell cycle and metastasis in a ZEB1-dependent manner. So, we constructed a marker vector labeled with ZEB1 and transfected it into PCAT6 knockout OS cells (Figure 4A). As shown in Figure 4B–D, the decrease in the proliferation rate of PCAT6 knockout cells was ameliorated after ZEB1 overexpression. Consistent with these data, the downregulation of PCAT6 expression resulted in cell cycle arrest following overexpression of ZEB1 in OS cells (Figure 4E and F). Moreover, the ZEB1 enhancement restored the invasion and metastasis of MG-63 and U2OS cells via PCAT6 expression knockout (Figure 4G and H). Therefore, PCAT6 regulates ZEB1 expression in a transcription-dependent manner, and PCAT6 functions as oncogene by positively regulating ZEB1 in OS.
MiR-143-3p/ZEB1 Axis is Responsible for PCAT6-Mediated Tumor-Promoting Effect in OS
Evidence suggests that lncRNAs located in the cytoplasm may act as competing endogenous RNA (ceRNAs) for specific small RNAs, thereby releasing the corresponding small RNA-targeted transcripts.22,23 To explore the mechanism underlying the role of PCAT6 in regulating ZEB1 expression, we examined the subcellular distribution of PCAT6 in MG-63 and U2OS cells. As shown in Figure 5A, PCAT6 was mainly located in the cytoplasm of OS cells. The binding sites of PCAT6 and small RNA were predicted with starBase V2.0. In several small RNAs, PCAT6 and miR-143-3p had highly predicted binding sites, as confirmed with the luciferase reporter analysis (Figure 5B). RIP analysis also showed that both PCAT6 and miR-143-3p were preferentially enriched in small RNA containing Ago2, rather than IgG, suggesting that PCAT6 and miR-143-3p had highly predictable binding sites. The same RNA-induced silencing complex (RISC) (Figure 5C). Therefore, these data suggest that PCAT6 acts as a molecular sponge in OS cells. As a tumour suppressor, miR-143-3p is known to be involved in various types of tumours. Here, we also found that the level of miR-143-3p was down-regulated in OS tissues than that in normal bone tissues (Supplemental Figure 1A and B). Furthermore, the expression level of miR-514306-3p was negatively correlated with the level of PCAT6 in OS tissues (Supplemental Figure 1C). Thus, these results indicate that PCAT6 targeted and negatively regulated miR-143-3p in OS.
A previous study showed that ZEB1 was the target of miR-143-3p.17 Consistently, we also found that miR-143-3p inhibitors prevented the suppression of ZEB1 protein expression in OS cells in response to PCAT6 knockout (Figure 5D and E). Meanwhile, the antineoplastic effect of PCAT6 silencing could be partially offset after treatment with miR-143-3p inhibitor (Figure 5F–H and Supplemental Figure 1D–F). In conclusion, the carcinogenic effect of PCAT6 in OS may be partially dependent on the regulation of the miR-143-3p/ZEB1 axis.
Discussion
PCAT6 is a newly discovered lncRNA, which has been proved to be related to the development and progress of several malignant tumours.24,25 Here, we found that the expression level of PCAT6 was significantly higher in patients with OS than in the corresponding normal bone tissues. It is worth noting that high PCAT6 levels were closely related to OS, tumour size, Enneking stage, distant metastasis, and shortened overall survival. PCAT6 exerted carcinogenic effects in OS that were partly dependent on the miR-143-3p/ZEB1 axis.
Studies have shown that lncRNAs as oncogenes or tumour suppressors are associated with cancer progression.26,27 For instance, the lncRNA, LSINCT5 acts as an oncogene26 by increasing the inhibitory effect of EZH2-induced allophycocyanin expression in OS. Cai et al found that the lncRNA NBR2 inhibited the epithelial–mesenchymal transition (EMT) of OS cells by regulating Notch1 signalling.28 Here, the results of functional analyses confirmed that PCAT6 silencing significantly inhibited the proliferation of OS cells. We also found that the knockout of PCAT6 expression resulted in the blockade of cells in the G0/G1 cell cycle, suggesting that the PCAT6-mediated growth of OS was related to cell cycle. In addition, our data suggest that PCAT6 knockout significantly reduced the migration and invasion of OS cells. The key role of PCAT6 in the malignant phenotype of OS cells was identified in vitro and the effect of PCAT6 on the growth and metastasis of OS cells was confirmed.
To explore the mechanism underlying PCAT6-mediated OS progression, we used ITRAQ method combined with MS analysis. The change in ZEB1 expression level was related to PCAT6 expression. ZEB1 is a transcription factor that regulates cell differentiation and performs tissue-specific functions. ZEB1 promotes the growth and migration of many cancer cells.29,30 Previous reports have shown that the ZEB1-induced EMT contributes to the early pathogenesis and metastasis of lung cancer.31 Consistent with this result, we found that PCAT6 enhanced the expression of ZEB1 in a transcription-independent manner. We also found that the enhanced expression of ZEB1 promoted the malignant phenotype of OS cells.
Accumulated evidences have suggested a new regulatory mechanism between lncRNAs and miRNAs.32 LncRNAs act as sponges of small RNAs and eliminate their inhibitory effects in many cancers. For instance, the lncRNA PVT1 acts as a molecular sponge regulates the expression miR-519d-3p33 and promotes the development of laryngeal cancer. Huang et al reported that lncRNA near 1 promotes pancreatic cancer progression by acting as a ceRNA of miR-506-3p.19 Here, we also found that PCAT6 functioned as a sponge of miR-143-3p, thereby acting as an anti-tumour gene17,34 in OS. Furthermore, we investigated whether miR-143-3p was involved in the antineoplastic effect of PCAT6 silencing on the growth and metastasis of OS. Rescue experiments showed that miR-143-3p inhibition contributed to the carcinogenicity of PCAT6 in OS cells. In addition, we found that miR-149-5p expression inhibition could restore the expression of ZEB1 protein in OS cells stably knocked out for PCAT6 expression. Moreover, the overexpression of ZEB2 in OS cells could eliminate the anti-tumour effects of PCAT6 silencing. Therefore, these data indicate that PCAT6 accelerates OS progression by regulating the miR-143-3p/ZEB1 pathway.
In summary, our data reveal that PCAT6 plays an important tumour-promoting role by regulating the miR-143-3p/ZEB1 axis in OS. These results demonstrate that PCAT6 may serve as a potential biomarker for the diagnosis of OS and act as a therapeutic target for patients with OS.
Disclosure
The authors declare no conflicts of interest for this work.
References
1. Mirabello L, Troisi RJ, Savage SA. Osteosarcoma incidence and survival rates from 1973 to 2004: data from the surveillance, epidemiology, and end results program. Cancer. 2009;115(7):1531–1543. doi:10.1002/cncr.24121
2. Isakoff MS, Bielack SS, Meltzer P, Gorlick R. Osteosarcoma: current treatment and a collaborative pathway to success. J Clin Oncol. 2015;33(27):3029–3035. doi:10.1200/JCO.2014.59.4895
3. Nie Z, Peng H. Osteosarcoma in patients below 25 years of age: an observational study of incidence, metastasis, treatment and outcomes. Oncol Lett. 2018;16(5):6502–6514. doi:10.3892/ol.2018.9453
4. Broadhead ML, Clark JC, Myers DE, Dass CR, Choong PF. The molecular pathogenesis of osteosarcoma: a review. Sarcoma. 2011;2011:959248. doi:10.1155/2011/959248
5. Fatica A, Bozzoni I. Long non-coding RNAs: new players in cell differentiation and development. Nat Rev Genet. 2014;15(1):7–21. doi:10.1038/nrg3606
6. Mercer TR, Dinger ME, Mattick JS. Long non-coding RNAs: insights into functions. Nat Rev Genet. 2009;10(3):155–159. doi:10.1038/nrg2521
7. Gooding AJ, Zhang B, Gunawardane L, Beard A, Valadkhan S, Schiemann WP. The lncRNA BORG facilitates the survival and chemoresistance of triple-negative breast cancers. Oncogene. 2018.
8. Gooding AJ, Zhang B, Gunawardane L, Beard A, Valadkhan S, Schiemann WP. The lncRNA BORG facilitates the survival and chemoresistance of triple-negative breast cancers. Oncogene. 2019;38(12):2020–2041. doi:10.1038/s41388-018-0586-4
9. Lorenzi L, Avila Cobos F, Decock A, et al. Long noncoding RNA expression profiling in cancer: challenges and opportunities. Genes Chromosomes Cancer. 2019;58(4):191–199. doi:10.1002/gcc.22709
10. Zhang S, Du L, Wang L, et al. Evaluation of serum exosomal LncRNA-based biomarker panel for diagnosis and recurrence prediction of bladder cancer. J Cell Mol Med. 2019;23(2):1396–1405. doi:10.1111/jcmm.14042
11. Liu ZB, Tang C, Jin X, Liu SH, Pi W. Increased expression of lncRNA SNHG12 predicts a poor prognosis of nasopharyngeal carcinoma and regulates cell proliferation and metastasis by modulating Notch signal pathway. Cancer Biomark. 2018;23(4):603–613. doi:10.3233/CBM-181873
12. Du Z, Fei T, Verhaak RG, et al. Integrative genomic analyses reveal clinically relevant long noncoding RNAs in human cancer. Nat Struct Mol Biol. 2013;20(7):908–913. doi:10.1038/nsmb.2591
13. Cui LH, Xu HR, Yang W, Yu LJ. lncRNA PCAT6 promotes non-small cell lung cancer cell proliferation, migration and invasion through regulating miR-330-5p. Onco Targets Ther. 2018;11:7715–7724. doi:10.2147/OTT.S178597
14. Wan L, Zhang L, Fan K, Cheng ZX, Sun QC, Wang JJ. Knockdown of long noncoding RNA PCAT6 inhibits proliferation and invasion in lung cancer cells. Oncol Res. 2016;24(3):161–170. doi:10.3727/096504016X14618564639178
15. Xu Y, Sun JY, Jin YF, Yu H. PCAT6 participates in the development of gastric cancer through endogenously competition with microRNA-30. Eur Rev Med Pharmacol Sci. 2018;22(16):5206–5213. doi:10.26355/eurrev_201808_15718
16. He Z, Yi J, Liu X, et al. MiR-143-3p functions as a tumor suppressor by regulating cell proliferation, invasion and epithelial-mesenchymal transition by targeting QKI-5 in esophageal squamous cell carcinoma. Mol Cancer. 2016;15(1):51. doi:10.1186/s12943-016-0533-3
17. Sun X, Dai G, Yu L, Hu Q, Chen J, Guo W. miR-143-3p inhibits the proliferation, migration and invasion in osteosarcoma by targeting FOSL2. Sci Rep. 2018;8(1):606. doi:10.1038/s41598-017-18739-3
18. Chen L, Yao H, Wang K, Liu X. Long non-coding RNA MALAT1 regulates ZEB1 expression by sponging miR-143-3p and promotes hepatocellular carcinoma progression. J Cell Biochem. 2017;118(12):4836–4843. doi:10.1002/jcb.26158
19. Huang B, Liu C, Wu Q, et al. Long non-coding RNA NEAT1 facilitates pancreatic cancer progression through negative modulation of miR-506-3p. Biochem Biophys Res Commun. 2017;482(4):828–834. doi:10.1016/j.bbrc.2016.11.120
20. Huang J, Deng G, Liu T, Chen W, Zhou Y. Long noncoding RNA PCAT-1 acts as an oncogene in osteosarcoma by reducing p21 levels. Biochem Biophys Res Commun. 2018;495(4):2622–2629. doi:10.1016/j.bbrc.2017.12.157
21. Wang Y, Zhang L, Zheng X, et al. Long non-coding RNA LINC00161 sensitises osteosarcoma cells to cisplatin-induced apoptosis by regulating the miR-645-IFIT2 axis. Cancer Lett. 2016;382(2):137–146. doi:10.1016/j.canlet.2016.08.024
22. Cao C, Zhang T, Zhang D, et al. The long non-coding RNA, SNHG6-003, functions as a competing endogenous RNA to promote the progression of hepatocellular carcinoma. Oncogene. 2017;36(8):1112–1122. doi:10.1038/onc.2016.278
23. Yan K, Tian J, Shi W, Xia H, Zhu Y. LncRNA SNHG6 is associated with poor prognosis of gastric cancer and promotes cell proliferation and EMT through epigenetically silencing p27 and sponging miR-101-3p. Cell Physiol Biochem. 2017;42(3):999–1012. doi:10.1159/000478682
24. Wan L, Zhang L, Fan K, Wang JJ. Diagnostic significance of circulating long noncoding RNA PCAT6 in patients with non-small cell lung cancer. Onco Targets Ther. 2017;10:5695–5702. doi:10.2147/OTT.S149314
25. Shi X, Liu Z, Liu Z, et al. Long noncoding RNA PCAT6 functions as an oncogene by binding to EZH2 and suppressing LATS2 in non-small-cell lung cancer. EBioMedicine. 2018;37:177–187. doi:10.1016/j.ebiom.2018.10.004
26. Kong D, Li C, Yang Q, Wei B, Wang L, Peng C. Long noncoding RNA LSINCT5 acts as an oncogene via increasing EZH2-induced inhibition of APC expression in osteosarcoma. Biochem Biophys Res Commun. 2018;507(1–4):193–197. doi:10.1016/j.bbrc.2018.11.005
27. Sui Y, Han Y, Zhao X, Li D, Li G. Long non-coding RNA GClnc1 promotes tumorigenesis in osteosarcoma by inhibiting p53 signaling. Biochem Biophys Res Commun. 2018;507(1–4):36–42. doi:10.1016/j.bbrc.2018.10.135
28. Cai W, Wu B, Li Z, et al. LncRNA NBR2 inhibits epithelial-mesenchymal transition by regulating Notch1 signaling in osteosarcoma cells. J Cell Biochem. 2018.
29. Krebs AM, Mitschke J, Lasierra Losada M, et al. The EMT-activator Zeb1 is a key factor for cell plasticity and promotes metastasis in pancreatic cancer. Nat Cell Biol. 2017;19(5):518–529. doi:10.1038/ncb3513
30. Hanrahan K, O’Neill A, Prencipe M, et al. The role of epithelial-mesenchymal transition drivers ZEB1 and ZEB2 in mediating docetaxel-resistant prostate cancer. Mol Oncol. 2017;11(3):251–265. doi:10.1002/1878-0261.12030
31. Peng DH, Ungewiss C, Tong P, et al. ZEB1 induces LOXL2-mediated collagen stabilization and deposition in the extracellular matrix to drive lung cancer invasion and metastasis. Oncogene. 2017;36(14):1925–1938. doi:10.1038/onc.2016.358
32. Yang C, Wu D, Gao L, et al. Competing endogenous RNA networks in human cancer: hypothesis, validation, and perspectives. Oncotarget. 2016;7(12):13479–13490. doi:10.18632/oncotarget.7266
33. Zheng X, Zhao K, Liu T, Liu L, Zhou C, Xu M. Long noncoding RNA PVT1 promotes laryngeal squamous cell carcinoma development by acting as a molecular sponge to regulate miR-519d-3p. J Cell Biochem. 2019;120(3):3911–3921. doi:10.1002/jcb.27673
34. Jiang X, Wang W, Yang Y, et al. Identification of circulating microRNA signatures as potential noninvasive biomarkers for prediction and prognosis of lymph node metastasis in gastric cancer. Oncotarget. 2017;8(39):65132–65142. doi:10.18632/oncotarget.17789
 © 2020 The Author(s). This work is published and licensed by Dove Medical Press Limited. The full terms of this license are available at https://www.dovepress.com/terms.php and incorporate the Creative Commons Attribution - Non Commercial (unported, v3.0) License.
By accessing the work you hereby accept the Terms. Non-commercial uses of the work are permitted without any further permission from Dove Medical Press Limited, provided the work is properly attributed. For permission for commercial use of this work, please see paragraphs 4.2 and 5 of our Terms.
© 2020 The Author(s). This work is published and licensed by Dove Medical Press Limited. The full terms of this license are available at https://www.dovepress.com/terms.php and incorporate the Creative Commons Attribution - Non Commercial (unported, v3.0) License.
By accessing the work you hereby accept the Terms. Non-commercial uses of the work are permitted without any further permission from Dove Medical Press Limited, provided the work is properly attributed. For permission for commercial use of this work, please see paragraphs 4.2 and 5 of our Terms.