Back to Journals » OncoTargets and Therapy » Volume 10
Long noncoding RNA NBAT-1 suppresses tumorigenesis and predicts favorable prognosis in ovarian cancer
Authors Yan CS, Jiang Y, Wan YC, Zhang L, Liu JH, Zhou SL, Cheng WJ
Received 14 October 2016
Accepted for publication 3 December 2016
Published 6 April 2017 Volume 2017:10 Pages 1993—2002
DOI https://doi.org/10.2147/OTT.S124645
Checked for plagiarism Yes
Review by Single anonymous peer review
Peer reviewer comments 3
Editor who approved publication: Dr Samir Farghaly
Changsheng Yan,1 Yi Jiang,1 Yicong Wan,1 Lin Zhang,2 Jinhui Liu,1 Shulin Zhou,1 Wenjun Cheng1
1Department of Gynecology, The First Affiliated Hospital of Nanjing Medical University, 2Department of Obstetrics and Gynecology, Zhongda Hospital Affiliated to Southeast University, Medical School, Southeast University, Nanjing, Jiangsu, People’s Republic of China
Abstract: Long noncoding RNA (lncRNA) has been proven to be involved in many biological processes in ovarian cancer (OC). However, the mechanism still remains unknown. In this study, we screened significantly downregulated NBAT-1, which has been proven to play a significant role in breast cancer, clear cell renal cell carcinoma, and neuroblastoma, but not in OC, in two independent datasets with relatively more samples (GSE18520 and GSE38666) from Gene Expression Omnibus. We found that lncRNA NBAT-1 was obviously downregulated in OC tissue compared to normal ovarian tissue (P<0.001) which was free of OC, and the detected levels of NBAT-1 were associated with the International Federation of Gynecology and Obstetrics stage and tumor size guidelines. Moreover, it has been shown that lower levels of NBAT-1 predict poor outcomes of OC. In order to investigate the functional role of NBAT-1, pcDNA-NBAT-1 and empty vector were transfected into TOV112D and OVCAR-3 cell lines. Overexpressed NBAT-1 significantly inhibited cell proliferation, invasion, and migration in both TOV112D and OVCAR-3 cell lines. Finally, Western blot assay indicated that NBAT-1 may exert its function by targeting the ERK1/2 and AKT signaling pathways. In addition, tumor formation growth assay showed that overexpressed NBAT-1 significantly suppresses tumor growth in vivo. In conclusion, our study suggests that NBAT-1 acts as an anti-oncogene in the development of OC.
Keywords: ovarian cancer, lncRNA NBAT-1, tumorigenesis, prognosis
Introduction
Ovarian cancer (OC), the most fatal gynecological cancer, is a common cause of cancer-related deaths in women worldwide.1–3 Despite developments in surgery and chemotherapy, the overall 5-year survival rate in OC patients remains only 30%.4 Patients often have an extremely poor outcome because of late clinical symptom presentation and rapid progression of the disease. In addition, this kind of malignancy usually implants extensively in the abdominal cavity.5 In order to develop better preventive and diagnostic approaches, as well as more effective treatment methods, a deep understanding of the molecular mechanisms implicated in the complex process of ovarian carcinogenesis is critical.
In the light of recent transcriptomic studies, it has become apparent that the majority of the transcribed genome is noncoding. Among these, there is a kind of noncoding RNA called long noncoding RNA (lncRNA, >200 bp in length), which has been proven to regulate many biological processes from nuclear organization to epigenetic modification of posttranscriptional regulation and RNA splicing,6,7 especially in cancers.
In previous studies, many lncRNAs have been reported to play crucial roles in epithelial OC (EOC). For example, HOTAIR was reported to promote proliferation, migration, and invasion in OC through the regulation of PIK3R3.8 HOST2 was proven to regulate biological behaviors of OC by mediating microRNA let-7b.9 Therefore, identification of OC-related lncRNA is necessary to understanding the progression and molecular mechanisms in order to establish better OC treatment methods.
NBAT-1 (also known as CASC14), transcribed from the intron of chromosome 6p22, was originally found to be associated with the progression and prognosis of neuroblastoma. A previous study predicted that the chromosome 6p22 locus is associated with clinically aggressive neuroblastoma.10 NBAT-1 also has been reported to regulate cell proliferation and neuronal differentiation and has a tight association with clinical outcome in neuroblastoma.11 Moreover, NBAT-1 has been proved to play an important role in breast cancer and clear cell renal cell carcinoma.12,13 However, the function of NBAT-1 still remains unknown in OC. The objective of our study was to detect the biological behaviors and molecular mechanisms of NBAT-1 in OC in order to seek a potential therapeutic target for OC patients.
Materials and methods
Tissue samples
A total of 86 patients were involved in this study at the First Affiliated Hospital of Nanjing Medical University (Jiangsu, China); of these patients, 57 underwent resection of the primary EOC, while the rest underwent resection of a normal ovary, which was free of OC, between 2011 and 2015. The study was approved by the Research Ethics Committee of Nanjing Medical University, and written informed consent was obtained from all patients. Tumor samples and normal tissues were rapidly frozen in liquid nitrogen and kept at −80°C until used. The tumor stage and grade was consistent with the International Federation of Gynecology and Obstetrics (FIGO) guidelines. Complete follow-up information was obtained for each patient until 2016. The survival time of each patient was counted from the day of the first operation to death or the last day of follow-up. The clinicopathological features of all patients are listed in Table 1.
Cell lines
Five human OC cell lines (A2780, TOV112D, HO-8910, OVCAR-3, and SKOV3) were obtained from the Chinese Type Culture Collection, Chinese Academy of Sciences, and were cultured in Dulbecco’s Modified Eagle Medium (Thermo Fisher Scientific, Waltham, MA, USA), supplemented with 10% fetal bovine serum (Gibco), 100 U/mL penicillin sodium, and 100 mg/mL streptomycin sulfate, at 37°C in a humidified air atmosphere containing 5% CO2. Cells were used when they were in the logarithmic growth phase.
RNA isolation and quantitative real-time polymerase chain reaction (qRT-PCR)
Total RNA was extracted from tissues and cell lines using the TRIzol reagent (Thermo Fisher Scientific) according to the manufacturer’s instructions. The expression levels of NBAT-1 were detected by qRT-PCR using the SYBR® Green Master Mix (TaKaRa BIO INC, Otsu, Japan), with GAPDH as an internal control. The primers are as follows: lncRNA NBAT-1 5′-ATTTCTGCTCCTGGGTCTTAC-3′ and 5′-AGTGGCTTGTCTGTTAGAGTC-3′ and GAPDH 5′-CACCCACTCCTCCACCTTTG-3′ and 5′-CCACCACCCTGTTGCTGTAG-3′. The comparative Ct method was used for the quantification of the transcripts.
Plasmid construction and cell transfection
Human NBAT-1 cDNA was ligated into the pcDNA 3.1 vector. Plasmid vectors (pcDNA-NBAT-1 and empty vector) for transfection were prepared using the EndoFree® Maxi Plasmid Kit (Qiagen, Hilden, Germany) and transfected into TOV112D and OVCAR-3 cells using Lipofectamine® 2000 (Invitrogen) according to the manufacturer’s protocol. At 48 h post transfection, cells were harvested for qRT-PCR or Western blot analysis.
Cell proliferation assay
To detect cell growth ability, 3×103 cells were seeded in a 96-well plate after transfection with plasmid vectors (pcDNA-NBAT-1 and empty vector) for 48 h. Then, 10 μL of Cell Count Kit-8 (CCK-8) was added to each well and incubated at 37°C for 3 h. The proliferation of cells was assessed at 6, 24, 48, 72, 96, and 120 h. Absorbance values at 490 nm were detected by the microplate reader.
For colony formation assay, 400 cells/well were seeded in six-well plates after transfection with plasmid vectors (pcDNA-NBAT-1 and empty vector) for 48 h. Two weeks later, colonies were fixed with methanol and stained with 0.1% crystal violet. The colonies with the diameters of >1 mm were counted.
In vitro cell invasion migration assays
For cell migration and invasion assays, 24-well Transwell chambers with 8 μm pore size polycarbonate membrane were used (Corning Inc., Corning, NY, USA). Cells were planted on the top side of the membrane precoated with Matrigel (BD, Franklin Lakes, NJ, USA; without Matrigel for cell migration assay) and incubated for 24 h. Cells inside the upper chamber were obliterated with cotton swabs, while cells on the lower membrane surface were fixed and then stained with 0.5% crystal violet solution. Five fields were counted randomly in each well.
Western blot assay
Western blot assays were performed using the following primary antibodies: antihuman MMP-2 (1:1,000; Abcam, Cambridge, MA, USA), MMP-9 (1:500; Proteintech, Danvers, MA, USA), ERK1/2 (1:1,000; Cell Signaling Technology, Danvers, MA, USA), p-ERK1/2 (1:1,000; Cell Signaling Technology), AKT (1:1,000; Cell Signaling Technology), p-AKT (1:1,000; Cell Signaling Technology), and GAPDH (1:1,000; bioWORLD, Dublin, OH, USA). Stimulated cells were lysed with RIPA lysis buffer (50 mM Tris–HCl [pH 7.5], 150 mM NaCl, 1% Triton X-100, 0.5% Na-deoxycholate) containing 1% protease inhibitors (phenylmethanesulfonyl fluoride); cell protein lysates were separated on 10% sodium dodecyl sulfate polyacrylamide gel electrophoresis gels and transferred to polyvinylidene fluoride membranes. The membranes were incubated with specific antibodies. Finally, electrochemiluminescence chromogenic substrates used in our experiment were quantified by densitometry.
In vivo tumor growth assays
Four-week female athymic BALB/c nude mice were maintained under specific pathogen-free conditions and manipulated according to protocols approved by the Shanghai Medical Animal Care Commission. pcDNA-NBAT-1 and empty vector stably transfected OVCAR-3 cells were harvested. For tumor formation assay, 107 cells were subcutaneously injected into a single side of each mouse. Tumor growth was examined every 3 days, and tumor volumes were calculated using the equation V = 0.5 × D × d2 (V, volume; D, longitudinal diameter; d, latitudinal diameter). This study was carried out in strict accordance with the recommendations in the Guide for the Care and Use of Laboratory Animals of the National Institutes of Health. The protocol was approved by the Committee on the Ethics of Animal Experiments of the Nanjing Medical University.
Statistical analysis
All statistical analyses in our experiment were performed using the SPSS version 20.0 software system (IBM, Armonk, NY, USA). Data are shown as mean ± standard error of mean (SEM). The differences between groups were analyzed by the Student’s t-test, Wilcoxon test, or χ2 test. The Kaplan–Meier method was performed for patients’ overall survival analysis. All experiments were run in triplicate. P<0.05 was considered statistically significant difference (*P<0.05; **P<0.01; ***P<0.001).
Results
Expression of NBAT-1 is downregulated, and correlation with clinicopathological factors in OC
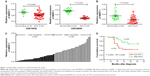
To detect the aberrant lncRNAs in EOC, we analyzed the microarray data from Gene Expression Omnibus (GEO) datasets and found that NBAT-1 was downregulated in both GSE18520 and GSE38666 (Figure 1A). Furthermore, the level of NBAT-1 was detected in 57 EOC tissues and 29 normal tissues by qRT-PCR and normalized to GAPDH. The results showed that NBAT-1 expression was significantly downregulated compared to normal counterparts (P<0.001; Figure 1B). According to the median ratio of relative NBAT-1 expression in tumor tissues (0.0152), the 57 EOC patients were classified into two groups: the relative low group (n=29, NBAT-1 expression ratio ≤ median ratio) and the relative high group (n=28, NBAT-1 expression ratio > median ratio; Figure 1C). The association between NBAT-1 expression and clinicopathological parameters was examined. As listed in Table 1, the NBAT-1 expression level was significantly associated with tumor size (P=0.024) and FIGO stage (P=0.029). However, there is no relation between NBAT-1 expression level and other parameters, such as age (P=0.357), histologic grade (P=0.234), and histological subtype (P=0.331) in OC.
Downregulated NBAT-1 predicts poor prognosis of EOC
The Kaplan–Meier survival analysis and log-rank test were performed to detect the correlation between NBAT-1 expression and EOC patients’ outcome. As shown in Figure 1D, patients with low levels of NBAT-1 expression have a shorter OS (months after diagnosis) than high-level groups (P=0.0463, hazard ratio =2.6260).
Modulation of NBAT-1 expression in OC cells
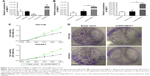
To detect the NBAT-1 expression level in diverse EOC cell lines, qRT-PCR was performed (Figure 2A). Then, NBAT-1 plasmid and plasmid-negative control was transfected into TOV112D and OVCAR-3 cell lines, which have lower expressions of NBAT-1 compared to other cell lines. Finally, qRT-PCR assays revealed that NBAT-1 expression was significantly upregulated in these two cell lines (Figure 2B).
Overexpression of NBAT-1 inhibits cell proliferation
In order to examine the proliferative ability of NBAT-1, CCK-8 assay was performed. As shown in Figure 2C, the NBAT-1-overexpressed group significantly inhibited cell proliferation in both TOV112D and OVCAR-3 cell lines compared to the empty vector group. Furthermore, colony formation was performed, and the result similarly indicates that clonogenic survival was significantly decreased in OVCAR-3 and TOV112D cell lines transfected with plasmid pcDNA-NBAT-1 group compared to the empty vector group (Figure 2D). Taken together, these results suggest that NBAT-1 is associated with proliferative ability in OVCAR-3 and TOV112D cells.
Overexpression of NBAT-1 suppresses OC cell migration and invasion
To investigate the roles of NABT-1 on the migration and invasion of OVCAR-3 and TOV112D cell lines, Transwell assays were performed to determine the migratory and invasive abilities of cells after the transfection of plasmid vectors (pcDNA-NBAT-1 and empty vector). Transwell migration assay revealed that the migration capacity was significantly suppressed in OVCAR-3 and TOV112D cell lines when plasmid pcDNA-NBAT-1 was transfected (Figure 3A and B). The invasion assay indicated that the cell numbers of invading cells were obviously lower in the plasmid pcDNA-NBAT-1 group than in the plasmid empty vector group (Figure 3A and B). These findings showed that NBAT-1 plays an important role in the migration and invasion capacity in both OVCAR-3 and TOV112D cell lines.
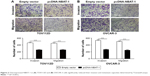  | Figure 3 Overexpressed NBAT-1 in (A) TOV112D and (B) OVCAR-3 cells significantly reduced their invasion and metastasis capacities determined by Transwell assays. |
Mechanisms of lncRNA NBAT-1 exert its function
In order to analyze the downstream genes of lncRNA NBAT-1, Western blot assays were performed. Consistent with functional characterization in vitro, the overexpression of NBAT-1 leads to signifcantly metastasis-related factors (MMP-2 and MMP-9) downregulated both in OVCAR-3 and TOV112D cell lines compared to the empty vector group (Figure 4A and B). As reported previously, the ERK1/2 and AKT signal pathways are usually activated in OC.14,15 As in previously published articles, our study showed that p-ERK1/2 and p-AKT protein expressions were reduced in the pcDNA-NBAT-1 group compared to the empty vector group in both TOV112D and OVCAR-3 cell lines. However, ERK1/2 and AKT protein expressions were the same, which show that lncRNA NBAT-1 may regulate tumor progress through the inhibition of the ERK1/2 and AKT signaling pathways (Figure 4A and B).
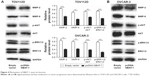  | Figure 4 Mechanisms of NBAT-1 exert its function. |
Overexpressed NBAT-1 inhibits OC cell proliferation in vivo
To detect whether NBAT-1 affects tumor growth in vivo, OVCAR-3 cells that were stably transfected with pcDNA-NBAT-1 and empty vector were inoculated into nude mice. All mice develop xenograft tumors at the injection site. Tumor growth in the pcDNA-NBAT-1 group was significantly slower than that in the empty vector group at the 21st day after injection (Figure 5A and B). Tumor weight in the pcDNA-NBAT-1 group was lower than that in the empty vector group (Figure 5C). To detect the expression of NBAT-1 in tumor tissues, qRT-PCR analysis was performed. Results showed that the level of NBAT-1 in the pcDNA-NBAT-1 group was obviously higher than that in the empty vector group (Figure 5C). Hematoxylin-eosin (H&E) staining showed the typical characteristics of tumor cells. The results of immunohistochemical staining showed that the proliferation index Ki-67 was significantly decreased in the pcDNA-NBAT-1 group (Figure 5D). Taken together, these results demonstrated that the overexpressed NBAT-1 could inhibit OC cell proliferation in vivo.
Discussion
In this study, we identified that NBAT-1, which has been proved to play an important role in breast cancer, clear cell renal cell carcinoma, and neuroblastoma11–13 but has not been reported in OC, was a significantly downregulated lncRNA in OC tissues. To further confirm the function of the lncRNA, we conducted a series of experiments.
In recent years, lncRNA has gained increasing attention as a novel class of molecules that demonstrate an important role in carcinogenesis.16,17 Previous studies have proven that the expression of lncRNAs is dysregulated in various human cancers, and these aberrant lncRNAs may play important roles during cancer progression, but the role of the majority of lncRNAs is still unknown.18–20 Until now, numerous lncRNAs have been reported to be abnormally expressed in OC, such as CCAT-1, NEAT-1, and CASC8. These lncRNAs are involved in various biological processes, including chemotherapy, proliferation, migration, and invasion.21–23
lncRNA NBAT-1 has been reported to play an important role in neuroblastoma and renal clear cell carcinoma,11,12 but its role in OC is still unclear. In this study, we confirmed that NBAT-1 is significantly downregulated in the EOC tissue compared with the normal ovarian tissue by analyzing the data from the GEO database and the tissues from patients. Then, the size of tumor and FIGO stage were associated with NBAT-1 level. The OS of the low-level group was shorter than the high-level group, indicating that lncRNA NBAT-1 plays an important role in EOC.
To further investigate the function of lncRNA NBAT-1 in EOC, two cell lines (OVCAR-3 and TOV112D) with lower NBAT-1 expression were chosen. Then, we transfected plasmid pcDNA-NBAT-1 and plasmid empty vector into these two OC cell lines. The results show that cell proliferation, invasion, and migration were significantly decreased. Thus, NBAT-1 plays an anti-oncogene role in EOC and represents a potential target for EOC treatment.
The exact mechanism by which lncRNA regulates tumor development and metastasis remains unclear. Possible RNA-targeting schemes include sequence-specific recognition (RNA–RNA), RNA–DNA hybrids, structure-mediated interactions, and protein-mediated interactions.24–26 For example, lncRNA HOTAIR acts as a sponge to mic-331-3p to regulate HER227 and lncRNA HULC enhances the epithelial–mesenchymal transition to promote the tumorigenesis and metastasis of hepatocellular carcinoma via the miR-200a-3p/ZEB1 signaling pathway.28 Many reports have demonstrated that lncRNA promotes tumor progression by regulating protein-coding genes. For instance, lncRNA AB073614 promotes tumor progression by regulating a series of invasion-associated proteins (MMP-2 and MMP-9)29 and other proteins. These findings prompted us to determine whether NBAT-1 promoted EOC progression and metastasis by regulating the expression of gene encoding proteins. As expected, the overexpression of NBAT-1 caused invasion and migration associated with proteins, such as MMP-2 and MMP-9, which were obviously downregulated in both TOV112D and OVCAR-3 cell lines. In addition, p-ERK1/2 and p-AKT were downregulated in NBAT-1-overexpressed cell lines with the same amount of ERK1/2 and AKT. This indicates that NBAT-1 regulates tumor progression through the ERK and AKT pathways. In addition, tumor formation in nude mice predicts that the overexpressed NBAT-1 could suppress OC cell proliferation in vivo.
Conclusion
This study suggests for the first time that lncRNA NBAT-1 acts as a functional anti-oncogene in EOC cell lines, and the downregulation of lncRNA NBAT-1 expression is closely associated with EOC development. Thus, these results indicate that lncRNA NBAT-1 may become a promising new candidate for EOC prognosis and therapy.
Disclosure
The authors report no conflicts of interest in this work.
References
Lalwani N, Prasad SR, Vikram R, Shanbhogue AK, Huettner PC, Fasih N. Histologic, molecular, and cytogenetic features of ovarian cancers: implications for diagnosis and treatment. Radiographics. 2011;31(3):625–646. | ||
Bast RC Jr, Hennessy B, Mills GB. The biology of ovarian cancer: new opportunities for translation. Nat Rev Cancer. 2009;9(6):415–428. | ||
Bowtell DD. The genesis and evolution of high-grade serous ovarian cancer. Nat Rev Cancer. 2010;10(11):803–808. | ||
Holmes D. Ovarian cancer: beyond resistance. Nature. 2015;527(7579):S217. | ||
Holmes D. The problem with platinum. Nature. 2015;527(7579):S218–S219. | ||
ENCODE Project Consortium. An integrated encyclopedia of DNA elements in the human genome. Nature. 2012;489(7414):57–74. | ||
Quinn JJ, Chang HY. Unique features of long non-coding RNA biogenesis and function. Nat Rev Genet. 2016;17(1):47–62. | ||
Dong L, Hui L. HOTAIR promotes proliferation, migration, and invasion of ovarian cancer SKOV3 cells through regulating PIK3R3. Med Sci Monit. 2016;22:325–331. | ||
Gao Y, Meng H, Liu S, et al. LncRNA-HOST2 regulates cell biological behaviors in epithelial ovarian cancer through a mechanism involving microRNA let-7b. Hum Mol Genet. 2015;24(3):841–852. | ||
Maris JM, Mosse YP, Bradfield JP, et al. Chromosome 6p22 locus associated with clinically aggressive neuroblastoma. N Engl J Med. 2008;358(24):2585–2593. | ||
Pandey GK, Mitra S, Subhash S, et al. The risk-associated long noncoding RNA NBAT-1 controls neuroblastoma progression by regulating cell proliferation and neuronal differentiation. Cancer Cell. 2014;26(5):722–737. | ||
Xue S, Li QW, Che JP, Guo Y, Yang FQ, Zheng JH. Decreased expression of long non-coding RNA NBAT-1 is associated with poor prognosis in patients with clear cell renal cell carcinoma. Int J Clin Exp Pathol. 2015;8(4):3765–3774. | ||
Hu P, Chu J, Wu Y, et al. NBAT1 suppresses breast cancer metastasis by regulating DKK1 via PRC2. Oncotarget. 2015;6(32):32410–32425. | ||
Huang WL, Li Z, Lin TY, Wang SW, Wu FJ, Luo CW. Thyrostimulin-TSHR signaling promotes the proliferation of NIH:OVCAR-3 ovarian cancer cells via trans-regulation of the EGFR pathway. Sci Rep. 2016;6:27471. | ||
Chen C, Chang YC, Lan MS, Breslin M. [Corrigendum] Leptin stimulates ovarian cancer cell growth and inhibits apoptosis by increasing cyclin D1 and Mcl-1 expression via the activation of the MEK/ERK1/2 and PI3K/Akt signaling pathways. Int J Oncol. 2016;49(2):847. | ||
Wang GY, Zhu YY, Zhang YQ. The functional role of long non-coding RNA in digestive system carcinomas. Bull Cancer. 2014;101(9):E27–E31. | ||
Gutschner T, Diederichs S. The hallmarks of cancer: a long non-coding RNA point of view. RNA Biol. 2012;9(6):703–719. | ||
Brunner AL, Beck AH, Edris B, et al. Transcriptional profiling of long non-coding RNAs and novel transcribed regions across a diverse panel of archived human cancers. Genome Biol. 2012;13(8):R75. | ||
Tano K, Mizuno R, Okada T, et al. MALAT-1 enhances cell motility of lung adenocarcinoma cells by influencing the expression of motility-related genes. FEBS Lett. 2010;584(22):4575–4580. | ||
Gupta RA, Shah N, Wang KC, et al. Long non-coding RNA HOTAIR reprograms chromatin state to promote cancer metastasis. Nature. 2010;464(7291):1071–1076. | ||
Huang S, Qing C, Huang Z, Zhu Y. The long non-coding RNA CCAT2 is up-regulated in ovarian cancer and associated with poor prognosis. Diagn Pathol. 2016;11(1):49. | ||
Jiang P, Wu X, Wang X, Huang W, Feng Q. NEAT1 upregulates EGCG-induced CTR1 to enhance cisplatin sensitivity in lung cancer cells. Oncotarget. 2016;7(28):43337–43351. | ||
Hu L, Chen SH, Lv QL, et al. Clinical significance of long non-coding RNA CASC8 rs10505477 polymorphism in lung cancer susceptibility, platinum-based chemotherapy response, and toxicity. Int J Environ Res Public Health. 2016;13(6):E545. | ||
Cesana M, Cacchiarelli D, Legnini I, et al. A long noncoding RNA controls muscle differentiation by functioning as a competing endogenous RNA. Cell. 2011;147(2):358–369. | ||
Li K, Blum Y, Verma A, et al. A noncoding antisense RNA in tie-1 locus regulates tie-1 function in vivo. Blood. 2010;115(1):133–139. | ||
Martianov I, Ramadass A, Serra Barros A, Chow N, Akoulitchev A. Repression of the human dihydrofolate reductase gene by a non-coding interfering transcript. Nature. 2007;445(7128):666–670. | ||
Liu XH, Sun M, Nie FQ, et al. Lnc RNA HOTAIR functions as a competing endogenous RNA to regulate HER2 expression by sponging miR-331-3p in gastric cancer. Mol Cancer. 2014;13:92. | ||
Li SP, Xu HX, Yu Y, et al. LncRNA HULC enhances epithelial–mesenchymal transition to promote tumorigenesis and metastasis of hepatocellular carcinoma via the miR-200a-3p/ZEB1 signaling pathway. Oncotarget. 2016;7(27):42431–42446. | ||
Cheng Z, Guo J, Chen L, Luo N, Yang W, Qu X. A long noncoding RNA AB073614 promotes tumorigenesis and predicts poor prognosis in ovarian cancer. Oncotarget. 2015;6(28):25381–25389. |
 © 2017 The Author(s). This work is published and licensed by Dove Medical Press Limited. The full terms of this license are available at https://www.dovepress.com/terms.php and incorporate the Creative Commons Attribution - Non Commercial (unported, v3.0) License.
By accessing the work you hereby accept the Terms. Non-commercial uses of the work are permitted without any further permission from Dove Medical Press Limited, provided the work is properly attributed. For permission for commercial use of this work, please see paragraphs 4.2 and 5 of our Terms.
© 2017 The Author(s). This work is published and licensed by Dove Medical Press Limited. The full terms of this license are available at https://www.dovepress.com/terms.php and incorporate the Creative Commons Attribution - Non Commercial (unported, v3.0) License.
By accessing the work you hereby accept the Terms. Non-commercial uses of the work are permitted without any further permission from Dove Medical Press Limited, provided the work is properly attributed. For permission for commercial use of this work, please see paragraphs 4.2 and 5 of our Terms.