Back to Journals » OncoTargets and Therapy » Volume 12
Long noncoding RNA LINC00673 promotes the proliferation and metastasis of epithelial ovarian cancer by associating with opioid growth factor receptor
Authors Zheng T, Qiu J, Li C , Lin X, Tang X, Hua K
Received 23 March 2019
Accepted for publication 27 June 2019
Published 5 August 2019 Volume 2019:12 Pages 6145—6156
DOI https://doi.org/10.2147/OTT.S209784
Checked for plagiarism Yes
Review by Single anonymous peer review
Peer reviewer comments 2
Editor who approved publication: Dr Arseniy Yuzhalin
Tingting Zheng,*,1,2 Junjun Qiu,*,1,2 Chunbo Li,1,2 Xiaojing Lin,1,2 Xiaoyan Tang,1,2 Keqin Hua1,2
1Department of Gynecology, Obstetrics and Gynecology Hospital, Fudan University, Shanghai 200011, People’s Republic of China; 2Shanghai Key Laboratory of Female Reproductive Endocrine-Related Diseases, Obstetrics and Gynecology Hospital, Fudan University, Shanghai 200011, People’s Republic of China
*These authors contributed equally to this work
Purpose: The long noncoding RNA LINC00673 has emerged as an important regulator of cancer development and progression. However, the clinical significance and biological roles of LINC00673 in epithelial ovarian cancer (EOC) remain unclear. In this study, we aimed to explore the oncogenic roles and underlying molecular mechanisms of LINC00673 in EOC.
Patients and methods: The expression levels of LINC00673 in EOC tissues and cell lines were detected by quantitative reverse transcription polymerase chain reaction (qRT-PCR). Real-time cellular analysis (RTCA), flow cytometry, and transwell assays were conducted to investigate cell proliferation, apoptosis, migration and invasion in vitro. Subcutaneous transplanted tumors were established to explore the oncogenic role of LINC00673 in vivo. Differentially expressed genes were analyzed using transcriptome sequencing. Protein levels were determined by Western blot assays.
Results: LINC00673 was upregulated in EOC tissues and cell lines compared to their corresponding normal controls. High expression of LINC00673 was associated with advanced International Federation of Gynecology and Obstetrics (FIGO) stage, serous histological subtype, lymph node metastasis and poor prognosis in patients with EOC. LINC00673 was also identified as an independent prognostic factor for EOC. In addition, LINC00673 promoted cell migration, invasion and proliferation and inhibited cell apoptosis in vitro and induced tumor growth in vivo. Mechanistically, opioid growth factor receptor (OGFR) was found to be a potential downstream target gene that mediated the oncogenic effect of LINC00673 in EOC.
Conclusion: LINC00673 contributes to EOC proliferation and metastasis and may be a promising prognostic biomarker for EOC patients.
Keywords: LINC00673, proliferation, metastasis, OGFR, ovarian cancer
Introduction
Epithelial ovarian cancer (EOC) is the most lethal gynecological cancer and the fifth most common cause of cancer-related death among women.1 Up to 70% of EOC patients are diagnosed in advanced stages with abdominal metastasis. Despite aggressive treatment, the prognosis of EOC patients still remains poor, with a 5-year survival rate of only 29%, which is mainly due to tumor recurrence or metastasis.2 Thus, there is a strong need to reveal the molecular mechanisms underlying EOC progression to identify reliable prognostic markers for EOC patients.
As an increasing research focus, long noncoding RNAs (lncRNAs, >200 nucleotides in length) open up new possibilities for the prognosis of cancer and provide deep insights into the biological behaviors of malignancies. For example, PCA3, HULC, and HOTAIR have been respectively regarded as prognostic biomarkers for prostate cancer, hepatocellular carcinoma and breast cancer.3–5 MEG3, LincRNA-p21, LINC00460 have also been found to play important roles in the development of metastasis and proliferation in malignancies.6–8 Together, these studies highlight the important roles of lncRNAs in cancer progression and prognosis. Most recently, LINC00673, a new lncRNA identified to be oncogenic in non-small cell lung cancer (NSCLC), has become the focus of our attention. Thus far, no published studies have investigated the clinical significance or biological roles of LINC00673 in EOC.
In this study, we aimed to explore the expression pattern, prognostic value, biological roles and mechanisms of LINC00673 in EOC. We found that LINC00673 was significantly elevated in EOC tissues and that it might be a potential predictor of poor prognosis for EOC patients. Moreover, silencing of LINC00673 suppressed migration, invasion and cell growth both in vivo and in vitro. Additionally, OGFR could restore the LINC00673 silencing-induced inhibition of malignancies. Taken together, our study is the first to reveal that LINC00673 may serve as a noncoding oncogene in EOC progression and that it may represent a novel prognostic marker for EOC.
Materials and methods
Tissue samples
One hundred and thirty-one EOC specimens and twenty-four normal ovarian tissues (NC) were obtained from the Tissue Bank of the Obstetrics and Gynecology Hospital of Shanghai Fudan University between 2010 and 2013 with approval from the ethics committee. A diagnosis of EOC was determined histologically by two experienced pathologists. One hundred and thirty-one patients with EOC underwent standard therapeutic approaches, including staging surgery or cytoreductive surgery. Patients with EOC complicated with other gynecological tumors and patients with radiotherapy or chemotherapy before surgery were excluded. Twenty-four normal ovarian samples were obtained from hysteromyoma patients scheduled to undergo hysterectomy and oophorectomy. The patients’ clinicopathological data were collected from the available medical records. Follow-up data, including overall survival (OS) and progression-free survival (PFS), were obtained by reviewing outpatient charts or by telephone interviews. Written Informed consent was obtained from all of the patients, according to the principles of the Declaration of Helsinki.
Cell culture and transfection
EOC cell lines (HO8910, OVCAR3, A2780, and ES2) and a normal human ovarian epithelial cell line (HOSEpiC) were purchased from the American Type Culture Collection (ATCC, Manassas, VA, USA). All cell lines were maintained in RPMI 1640 (Gibco, Thermo Fisher Scientific, Carlsbad, CA, USA) with 10% fetal bovine serum (Gibco, Thermo Fisher Scientific, Carlsbad, CA, USA) and 100 units/ml penicillin-streptomycin (Gibco, Thermo Fisher Scientific, Carlsbad, CA, USA) and were cultured in a humidified 5% CO2 incubator at 37 °C. Cells were transfected with LINC00673-siRNAs (GenePharma, Shanghai, China) using Lipofectamine 3000 transfection reagent (Invitrogen, Carlsbad, CA) for transwell assays. The siRNAs sequences were 5′-GCCUUUGUACUCAGCAAUUTT-3ʹ (LINC00673-siRNA1) and 5′-CCUUGGAAUAGUAACUCUUTT-3′ (LINC00673-siRNA2). Cells were transfected with a recombinant LINC00673-shRNA lentivirus constructed by GenePharma (Shanghai, China) for proliferation assays, flow cytometry and animal experiments. The LINC00673-shRNA-lentivirus-clones were designated as KD, and LINC00673-negative control-lentivirus-clones were named NC. The shRNA sequence was CCGGGCCTTTGTACTCAGCAATTCTCGAGAATTGCTGAGTACAAAGGCTTTTT. Additionally, the lentivirus expressing the LINC00673 sequence (LINC00673-overexpression, OE) and the negative control lentivirus (Vector) were constructed by GenePharma (Shanghai, China). For rescue assays, the siRNAs sequences targeted at OGFR were 5′-CGCCAAACCUGAGUUUCUATT-3ʹ (OGFR-siRNA1) and 5′-GCUGUUUCAUUGAGGACAUTT-3ʹ (OGFR-siRNA2).
For lentiviral transduction, cells were seeded in six-well plates at a density of 105 cells per well and cultured overnight. The next day, the cells were incubated with 5 μg/mL polybrene (GenePharma, China) for 30 min before being exposed to 1 mL of medium containing the same number of units of lentiviral particles for 24 h. The culture medium was replaced with selective medium containing 1 μg/mL puromycin after 72 h of transfection, and the cells were cultured for at least 14 days. The puromycin-resistant cell clones were isolated, amplified in medium containing 0.5 μg/mL puromycin for seven days and transferred to normal medium.
RNA isolation, reverse transcription, and qRT-PCR
Total RNA was extracted using TRIzol reagent (Invitrogen, Carlsbad, CA) and was reverse transcribed into cDNA using a Prime-Script RT reagent kit (TAKARA, Dalian, China). LINC00673 expression levels were measured with qRT-PCR using the SYBR Green PCR Master Mix (TAKARA, Dalian, China) on the Prism 7900 system (Applied Biosystems, Foster City, CA, USA). GAPDH was used as an endogenous standard. The sequences of primers designed by Sangon Biotech (Shanghai, China) were as follows: LINC00673 forward: 5ʹ-TACCACACCCTTTCTTGCCC-3ʹ, LINC00673 reverse: 5ʹ-ACACTGGCCTCTTTACACGG-3ʹ, GAPDH forward: 5ʹ-GGGAAGGTGAAGGTCGGAGT-3ʹ, GAPDH reverse: 5ʹ-GGGGTCATTGATGGCAACA-3ʹ.
Cell migration and invasion assays
Cell migration was assayed by using a 24-well transwell chamber (Corning, NY, USA) with polycarbonate filters (8-μm pore size). Cell invasion was performed with a Matrigel invasion chamber (Corning, NY, USA). A suspension of transfected cells (1.2×105 cells for HO8910 and A2780, 8×104 cells for OVCAR3 in 200 μL of serum-free RPMI 1640) was placed in the upper chambers, and the lower chambers were filled with 500 μL of RPMI 1640 containing 10% FBS. After 48 h, the cells on the upper side of the filter were removed using cotton swabs. The migrated cells that remained on the bottom surface were fixed with paraformaldehyde (PFA, 3.5%, 30 min) and were then stained with 0.1% crystal violet for 10 min. The number of cells in five random fields of each chamber was imaged (magnification, ×200) and averaged. The assays were conducted in triplicate.
Proliferation assay
The effect of LINC00673 on cell proliferation was determined using real-time cellular analysis (RTCA). Twenty-four hours after transfection, cells (5×103 cells/well) were seeded in several E-Plate 16 dishes (ACEA Biosciences Inc, CA, USA) for proliferation assays. The plates were kept in the cell incubator at 37 °C with 5% CO2 for 4–6 days. The cell index and growth curves were automatically recorded on the xCELLigence RTCA System (ACEA Biosciences Inc, CA, USA).
Flow cytometry (FCM)
Cells were harvested by 0.25% trypsin without EDTA (Gibco, Thermo Fisher Scientific, Carlsbad, CA, USA) 48 h after transfection. For the apoptosis assay, 1×106 transfected cells were resuspended in 100 μL of binding buffer and labeled with 5 μL of Annexin V and 5 μL of 7-AAD according to the protocol using the PE Annexin V Apoptosis Detection Kit I (BD biosciences, CA, USA). Flow cytometry assays were performed to detect the cell apoptosis ratio by FACStation (FV500, Beckman Coulter, Brea, CA, USA).
In vivo experiment
Fourteen female athymic BALB/c nude mice (aged 4–5 weeks, weighing 14–16 g) were purchased from the Department of Laboratory Animals, Fudan University. They were housed in a pathogen-free animal facility and were randomly assigned to the NC or KD group. Both groups were inoculated subcutaneously under the right axilla on day 0 with 107 HO8910 cells (KD or NC). The size (length and width) of the resultant xenograft tumors was measured every three days, and tumor volume was determined using the following formula: volume (mm3) =1/2 (L×W×W). The tumor-bearing mice were euthanized four weeks after the first injection of tumor cells, and the tumor weight was subsequently recorded. All animal studies were approved by the Institutional Animal Care and Use Committee of Fudan University and were performed in accordance with the experimental protocols of the institutional Animal Care and Use Committee of Fudan University (Shanghai, China).
Transcriptome sequencing
Twenty-four hours after transfection with LINC00673-siRNA or NC-scramble, total RNA was extracted from HO8910 cells using TRIzol reagent (Invitrogen, CA, USA). Next, we conducted RNA-sequencing analysis using an Illumina HiSeq 4000 to compare the transcriptome profiles of LINC00673 knockdown and negative controls. Differentially regulated genes were determined, and Gene Ontology (GO) and KEGG pathway enrichment analyses were performed to unveil the biological functions of the molecular changes.
Western blot assays
Cells were rinsed twice with ice-cold PBS and lysed with RIPA buffer (0.2 mL/6-cm plate) containing protease inhibitors (Roche, CA, USA). Extracts were obtained by centrifugation at 12,000 g for 30 min at 4 °C. Samples (30 µg) of the extracts were subjected to SDS-PAGE and transferred onto PVDF membranes (Millipore, NY, USA). Membranes were incubated with the corresponding primary antibodies overnight at 4 °C, followed by incubation with an HRP-conjugated secondary antibody (Bioworld Technology, CA, USA). Then, the bound antibodies were detected using an ECL kit (Millipore, NY, USA). Quantification was performed using QuantityOne software (Bio-Rad, CA, USA). The antibodies were as follows: rabbit anti-human OGFR (Abcam, Cambridge, MA, USA; 1:1000); rabbit anti-human GAPDH (Millipore, NY, USA; 1:10000); and HRP-conjugated mouse anti-rabbit IgG (Bioworld Technology, CA, USA; 1:5000).
Statistical analysis
All data were analyzed by using SPSS 21.0. The Kaplan–Meier method was used to calculate survival, and the log-rank test was used to compare the differences between the two groups. Multivariate and univariate Cox regression analyses were performed to identify the independent factors related to EOC prognosis. The relationship between LINC00673 expression levels and clinical parameters was assessed by the Pearson chi-square test (n>5 in subgroups) or Fisher’s exact test (n<5 in subgroups). Quantitative data was presented as the means ± SD. Continuous data were analyzed by Student’s t-test (two-tailed). P-values less than 0.05 were considered statistically significant.
Results
LINC00673 expression is upregulated in EOC tissues and predicts poor prognosis in EOC patients
LINC00673 expression in EOC tissues was measured by qRT-PCR. The results showed that LINC00673 levels were significantly higher in EOC tissues compared with NC tissues (Figure 1A), suggesting that LINC00673 may be an oncogenic lncRNA involved in EOC development. The 131 EOC patients were divided into a high-LINC00673 group (n=66) and a low-LINC00673-group (n=65), using the median value of LINC00673 expression as the cut-off. High LINC00673 levels were found to be associated with advanced FIGO stage, lymph node metastasis and serous histological subtype (Table 1), indicating that LINC00673 positively correlates with a high degree of malignancy of EOC. Based on the available follow-up data, 110 patients were further studied for EOC prognosis (follow-up rate, 84.0%). Shorter PFS (P<0.05) and OS (P<0.01) were observed in EOC patients with higher LINC00673 expression than in those with lower LINC00673 expression by Kaplan–Meier survival analysis (Figure 1B and C). Moreover, univariate and multivariate Cox regression analyses indicated that FIGO stage (P=0.027) and LINC0067 expression (P=0.015) were independent prognostic factors for patients with EOC (Table 2). These findings collectively demonstrated that LINC00673 was significantly correlated with aggressive behaviors of EOC and might represent an independent prognostic marker.
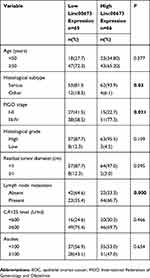 |
Table 1 Association of LINC00673 expression with clinicopathological variables in EOC patients |
 |
Table 2 Univariate and multivariate analyses of OS in 110 EOC patients |
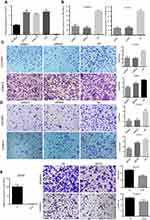
LINC00673 induces EOC cell migration and invasion in vitro
To explore the biological roles of LINC00673 in vitro, we detected the LINC00673 levels in EOC cell lines and found that LINC00673 was significantly upregulated in OVCAR3, HO8910, ES2 cells and downregulated in A2780 cells compared with HOSEpiC cells (Figure 2A).
Based on the clinical observations, we hypothesized that LINC00673 might contribute to EOC metastasis. To test our prediction, siRNAs were transfected to OVCAR3, HO8910 cells while LINC00673-overexpression lentivirus was transfected to A2780 cells to explore the function of LINC00673 in migration and invasion (Figure 2B and E). The migration assay results showed that the migratory potential of cells was significantly suppressed after LINC00673 silencing (Figure 2C). The Matrigel invasion assay results indicated that the knockdown of LINC00673 dramatically inhibited the invasive capacities of HO8910 and OVCAR3 cells (Figure 2D). In contrast, overexpression of LINC00673 in A2780 contributed to the cell migration and invasion (Figure 2E). Therefore, we concluded that elevated expression of LINC00673 promoted cell migration and invasion in EOC in vitro.
LINC00673 promotes EOC cell proliferation and inhibits cell apoptosis
To further determine whether LINC00673 is involved in cell proliferation and apoptosis, LINC00673-knockdown lentivirus cells (KD) or LINC00673-negative control lentivirus cells (NC) were developed in OVCAR3 and HO8910, and their silencing efficiencies were quantified by qRT-PCR. (Figure 3A). We performed RTCA and FCM tests to determine whether LINC00673 is involved in cell proliferation and apoptosis. Cell proliferation assays showed that LINC00673 silencing significantly inhibited the proliferation of HO8910 and OVCAR3 cells, while LINC00673 overexpression in A2780 cells contributed to cell growth (Figure 3B). In addition, we found that inhibition of LINC00673 caused marked increases in the percentages of apoptotic cells in the HO8910 and OVCAR3 cell lines, while upregulation of LINC00673 suppressed cell apoptosis in A2780 cells (Figure 3C). In summary, these results suggest that LINC00673 promotes proliferation and inhibits cell apoptosis in vitro.
LINC00673 knockdown inhibits tumor growth in vivo
To elucidate the effect of LINC00673 in vivo, xenograft tumors were established in nude mice by subcutaneous inoculation with KD-HO8910 or NC-HO8910 cells. As shown in Figure 3A, LINC00673 was effectively downregulated in the KD-HO8910 compared with the NC-HO8910. Tumor growth in the KD group was significantly slower than that in the NC group, and the growth curves of the two groups showed a widening gap over time (Figure 3D). After four weeks, the mice were sacrificed, and the tumors were removed. The knockdown efficiency of LINC00673 in vivo was verified by qRT-PCR (Figure 3G). We found that tumors in the KD group were substantially smaller than those in the NC group (Figure 3E and F). The tumor weight in the KD group was almost 65% less than that in the NC group (Figure 3H). To summarize, these results indicate that silencing of LINC00673 impairs tumor growth in vivo.
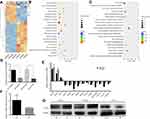
LINC00673 suppresses the transcription of OGFR in EOC
To further explore the molecular mechanism by which LINC00673 affects tumor proliferation and metastasis, we performed transcriptome sequencing analysis to compare the transcriptome profiles between LINC00673-KD and LINC00673-NC samples in HO8910 cells. The silencing efficiency was verified by qRT-PCR (Figure 4D). The results showed that 78 genes were markedly dysregulated (>1.5-fold change, 53 downregulated and 25 upregulated) after LINC00673 silencing, as shown in the heat map (Figure 4A). Based on KEGG database, the differentially expressed genes were found to enrich in MAPK signaling pathway, Ras signaling pathway and focal adhesion (Figure 4B), which were usually involved in cell proliferation and metastasis. The GO analysis showed that wound healing and epithelial cell proliferation changed significantly after LINC00673 knockdown (Figure 4C). These results collectively suggested that the dysregulated genes after LINC00673 silencing were associated with tumor growth and metastasis. Fourteen differential genes were further validated by qRT-PCR and Western blotting assays in both HO8910 and OVCAR3 cells and the results were consistent with transcriptome sequencing data. As shown in Figure 4E, knockdown of LINC00673 increased the expression of OGFR, RGS19, FICD and decreased the expression of CDC42, DCP1A, TAF12, GPAT3, PALM2, STX12, LMNBI, PPP1R14B, COL4A1, ELOVL1 and MTHFD2. Among those genes, OGFR was significantly upregulated both in LINC00673-KD cell lines and LINC00673-KD xenograft tumors (Figure 4E and F). Besides, the Western blotting assays revealed that OGFR was significantly upregulated in LINC00673-KD cells while was downregulated in LINC00673-OE cells compared to their corresponding controls (Figure 4G), which indicated that OGFR might be the key downstream mediator of LINC00673.
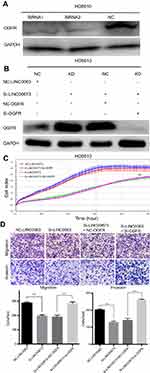
Knockdown of OGFR ablated the promotion effect of LINC00673 in cell proliferation, migration and invasion
To investigate whether OGFR mediated the role of LINC00673 in cell proliferation and metastasis, we used siRNAs to knockdown OGFR expression for further rescue experiments (Figure 5A). The results showed that OGFR was significantly decreased after co-transfection of si-OGFR and si-LINC00673 compared with that observed in cells treated with si-LINC00673 (Figure 5B). The RTCA and transwell assays demonstrated that downregulation of OGFR could restore the cell proliferation, migration and invasion that were inhibited by si-LINC00673 in the HO8910 cells (Figure 5C and D). Taken together, these data indicate that upregulation of LINC00673 promotes EOC proliferation and metastasis, at least in part by dysregulating OGFR as a downstream targeted gene.
Discussion
As a major component of noncoding RNAs, LncRNAs have emerged as critical players in tumorigenesis and cancer progression in recent years. Increasing evidence has indicated that LINC00673 might play important roles in a variety of malignancies.9–18 However, the potential functions and mechanistic details of LINC00673 in EOC remain unclear.
In this study, we found that LINC00673 was significantly upregulated in EOC tissues and cells. High expression of LINC00673 was associated with advanced FIGO stage, lymph node metastasis, serous histological subtype and poor prognosis. Univariate and multivariate Cox regression analyses showed that LINC00673 was an independent predictive marker for OS. These findings indicate that LINC00673 is closely associated with malignant features of EOC and could serve as a prognostic factor for EOC patients.
Previously, researchers found that upregulation of LINC00673 in many epithelial tumors induced cell proliferation, migration and invasion and contributed to epithelial-to-mesenchymal transition (EMT). In the present study, we conducted a series of experiments in EOC both in vitro and in vivo. The results showed that LINC00673 silencing significantly inhibited cell proliferation, induced apoptosis and decreased cell migration and invasion, while overexpression of LINC00673 showed the opposite results. In vivo, we found that LINC00673 knockdown led to dramatic decreases in the tumor growth rate and tumor volume. These results collectively suggest that LINC00673 is an important oncogenic factor in EOC and that its overexpression contributes to EOC proliferation and metastasis.
To date, our understanding of the molecular mechanisms by which lncRNAs act in tumor development and metastasis is far from adequate. It is clear that lncRNAs regulate tumor biological behaviors through various mechanisms, including regulating certain protein-coding genes. For instance, lncRNA THOR upregulates the expression of c-myc to promote tumor growth.19 HOTAIR regulates the expression of a series of epithelial-to-mesenchymal transition (EMT)-related genes to promote tumor metastasis.20 Inspired by these findings, we hypothesized that LINC00673 might dysregulate downstream coding genes to promote tumor progression. To this end, we conducted transcriptome sequencing tests to identify potential downstream target genes. Among the differential expressed genes, OGFR caught our attention. OGFR is known to be involved in cell proliferation in various cancers and the p21WAF1/CIP1 pathway.21–25 Zagon et al26 also found that the OGFR antagonist NTX could promote cell proliferation in 31 human cancer cell lines. These studies collectively demonstrated that the expression level of OGFR is an important regulator of cell proliferation. Our study showed that OGFR was significantly upregulated after LINC00673 knockdown. To further clarify the correlation between OGFR and LINC00673, we performed OGFR rescue experiment and found that knockdown of OGFR restored cell proliferation and metastasis that were inhibited by si-LINC00673 in HO8910 cells. Taken together, these data implied that LINC00673 regulated EOC proliferation and metastasis at least in part by dysregulating OGFR.
However, further investigations are needed to elucidate the regulatory mechanisms underlying the relationship between LINC00673 and OGFR. In addition, studies with larger samples are required to enhance the feasibility and reliability of LINC00673 as a prognostic biomarker for EOC.
Conclusion
We provided the first evidence for the oncogenic effect of LINC00673 in EOC, which is partly by dysregulating OGFR as a downstream targeted gene, and demonstrated that LINC00673 may be a promising prognostic biomarker for EOC patients.
Abbreviation list
EOC, epithelial ovarian cancer; OGFR, opioid growth factor receptor; RTCA, real-time cellular analysis; FCM, flow cytometry; FIGO, International Federation of Gynecology and Obstetrics; lncRNA, long noncoding RNA; NSCLC, non-small cell lung cancer; OS, overall survival; PFS, progression-free survival; EMT, epithelial-to-mesenchymal transition.
Ethics approval
This study was approved by the ethics committee of the Obstetrics and Gynecology Hospital of Fudan University (No. [2017]33)
Data availability
The datasets used and/or analyzed during the present study are available from the corresponding author on reasonable request.
Acknowledgments
This project was supported by funding from the National Natural Science Foundation of China (No.81571404), the Shanghai Science and Technology Innovation Foundation (No. 16411950500), the Research and Innovation Project of Shanghai Municipal Education Commission (No. 2019-01-07-00-07-E00050), the National Natural Science Foundation for Young Scholars of China (81502240), and the Shanghai Science and Technology Natural Science Foundation (No. 19ZR1406900).
Author contributions
All authors contributed to data analysis, drafting and revising the article, gave final approval of the version to be published, and agree to be accountable for all aspects of the work.
Disclosure
The authors report no conflicts of interest related to this work.
References
1. Jemal A, Bray F, Center MM, Ferlay J, Ward E, Forman D. Global cancer statistics. CA Cancer J Clin. 2011;61(2):69–90. doi:10.3322/caac.20107
2. Siegel RL, Miller KD, Jemal A. Cancer statistics, 2018. CA Cancer J Clin. 2018;68(1):7–30. doi:10.3322/caac.21442
3. Hessels D, Klein Gunnewiek JM, van Oort I, et al. DD3(PCA3)-based molecular urine analysis for the diagnosis of prostate cancer. Eur Urol. 2003;44(1):8–15. doi:10.1016/S0302-2838(03)00201-X
4. Panzitt K, Tschernatsch MM, Guelly C, et al. Characterization of HULC, a novel gene with striking up-regulation in hepatocellular carcinoma, as noncoding RNA. Gastroenterology. 2007;132(1):330–342. doi:10.1053/j.gastro.2006.08.026
5. Gupta RA, Shah N, Wang KC, et al. Long non-coding RNA HOTAIR reprograms chromatin state to promote cancer metastasis. Nature. 2010;464(7291):1071–1076. doi:10.1038/nature08940
6. Zhou Y, Zhang X, Klibanski A. MEG3 noncoding RNA: a tumor suppressor. J Mol Endocrinol. 2012;48(3):45–53. doi:10.1530/JME-12-0008
7. Chen S, Liang H, Yang H, et al. LincRNa-p21: function and mechanism in cancer. Med Oncol. 2017;34(5):98. doi:10.1007/s12032-017-0959-5
8. Lian Y, Yan C, Xu H, et al. A novel incRNA, LINC00460, affects cell proliferation and apoptosis by regulating KLF2 and CUL4A expression in colorectal cancer. Mol Ther Nucleic Acids. 2018;12:684–697. doi:10.1016/j.omtn.2018.06.012
9. Ba MC, Long H, Cui SZ, et al. Long noncoding RNA LINC00673 epigenetically suppresses KLF4 by interacting with EZH2 and DNMT1 in gastric cancer. Oncotarget. 2017;8(56):95542–95553. doi:10.18632/oncotarget.20980
10. Huang M, Hou J, Wang Y, et al. Long noncoding RNA LINC00673 Is activated by SP1 and exerts oncogenic properties by interacting with LSD1 and EZH2 in gastric cancer. Mol Ther. 2017;25(4):1014–1026. doi:10.1016/j.ymthe.2016.10.004
11. Lu W, Zhang H, Niu Y, et al. Erratum to: long non-coding RNA linc00673 regulated non-small cell lung cancer proliferation, migration, invasion and epithelial mesenchymal transition by sponging miR-150-5p. Mol Cancer. 2017;16(1):144. doi:10.1186/s12943-017-0716-6
12. Lu W, Zhang H, Niu Y, et al. Long non-coding RNA linc00673 regulated non-small cell lung cancer proliferation, migration, invasion and epithelial mesenchymal transition by sponging miR-150-5p. Mol Cancer. 2017;16(1):118. doi:10.1186/s12943-017-0685-9
13. Xia E, Bhandari A, Shen Y, Zhou X, Wang O. lncRNA LINC00673 induces proliferation, metastasis and epithelial-mesenchymal transition in thyroid carcinoma via Kruppel-like factor 2. Int J Oncol. 2018;53(5):1927–1938.
14. Xia E, Shen Y, Bhandari A, et al. Long non-coding RNA LINC00673 promotes breast cancer proliferation and metastasis through regulating B7-H6 and epithelial-mesenchymal transition. Am J Cancer Res. 2018;8(7):1273–1287.
15. Yu J, Liu Y, Gong Z, et al. Overexpression long non-coding RNA LINC00673 is associated with poor prognosis and promotes invasion and metastasis in tongue squamous cell carcinoma. Oncotarget. 2017;8(10):16621–16632.
16. Zhang F, Huang Y, Wang B, Zhong C, Liu X, Ding S. LINC00673 silencing inhibits cell migration and invasion by suppressing PI3K/AKT signaling in glioma. Neuroreport. 2018;29(9):718–722. doi:10.1097/WNR.0000000000001022
17. Zhang LG, Zhou XK, Zhou RJ, Lv HZ, Li WP. Long non-coding RNA LINC00673 promotes hepatocellular carcinoma progression and metastasis through negatively regulating miR-205. Am J Cancer Res. 2017;7(12):2536–2544.
18. Zheng J, Huang X, Tan W, et al. Pancreatic cancer risk variant in LINC00673 creates a miR-1231 binding site and interferes with PTPN11 degradation. Nat Genet. 2016;48(7):747–757. doi:10.1038/ng.3568
19. Shang Y. LncRNA THOR acts as a retinoblastoma promoter through enhancing the combination of c-myc mRNA and IGF2BP1 protein. Biomed Pharmacother. 2018;106:1243–1249. doi:10.1016/j.biopha.2018.07.052
20. Qiu JJ, Lin YY, Ye LC, et al. Overexpression of long non-coding RNA HOTAIR predicts poor patient prognosis and promotes tumor metastasis in epithelial ovarian cancer. Gynecol Oncol. 2014;134(1):121–128. doi:10.1016/j.ygyno.2014.03.556
21. Kim JY, Ahn HJ, Kim JK, Kim J, Lee SH, Chae HB. Morphine suppresses lung cancer cell proliferation through the interaction with opioid growth factor receptor: an in vitro and human lung tissue study. Anesth Analg. 2016;123(6):1429–1436. doi:10.1213/ANE.0000000000001293
22. Cheng F, McLaughlin PJ, Verderame MF, Zagon IS. The OGF-OGFr axis utilizes the p21 pathway to restrict progression of human pancreatic cancer. Mol Cancer. 2008;7:5. doi:10.1186/1476-4598-7-5
23. Cheng F, McLaughlin PJ, Verderame MF, Zagon IS. Dependence on nuclear localization signals of the opioid growth factor receptor in the regulation of cell proliferation. Exp Biol Med (Maywood). 2009;234(5):532–541. doi:10.3181/0901-RM-16
24. Cheng F, McLaughlin PJ, Verderame MF, Zagon IS. The OGF-OGFr axis utilizes the p16INK4a and p21WAF1/CIP1 pathways to restrict normal cell proliferation. Mol Biol Cell. 2009;20(1):319–327. doi:10.1091/mbc.e08-07-0681
25. Zagon IS, Verderame MF, McLaughlin PJ. The expression and function of the OGF-OGFr axis - a tonically active negative regulator of growth - in COS cells. Neuropeptides. 2003;37(5):290–297.
26. Zagon IS, Donahue RN, McLaughlin PJ. Opioid growth factor-opioid growth factor receptor axis is a physiological determinant of cell proliferation in diverse human cancers. Am J Physiol Regul Integr Comp Physiol. 2009;297(4):1154–1161. doi:10.1152/ajpregu.00414.2009
 © 2019 The Author(s). This work is published and licensed by Dove Medical Press Limited. The full terms of this license are available at https://www.dovepress.com/terms.php and incorporate the Creative Commons Attribution - Non Commercial (unported, v3.0) License.
By accessing the work you hereby accept the Terms. Non-commercial uses of the work are permitted without any further permission from Dove Medical Press Limited, provided the work is properly attributed. For permission for commercial use of this work, please see paragraphs 4.2 and 5 of our Terms.
© 2019 The Author(s). This work is published and licensed by Dove Medical Press Limited. The full terms of this license are available at https://www.dovepress.com/terms.php and incorporate the Creative Commons Attribution - Non Commercial (unported, v3.0) License.
By accessing the work you hereby accept the Terms. Non-commercial uses of the work are permitted without any further permission from Dove Medical Press Limited, provided the work is properly attributed. For permission for commercial use of this work, please see paragraphs 4.2 and 5 of our Terms.