Back to Journals » Cancer Management and Research » Volume 12
Long Noncoding RNA LINC00460 Modulates MMP-9 to Promote Cell Proliferation, Invasion and Apoptosis by Targeting miR-539 in Papillary Thyroid Cancer
Authors Zou X, Guo ZH, Li Q, Wang PS
Received 5 July 2019
Accepted for publication 18 November 2019
Published 9 January 2020 Volume 2020:12 Pages 199—207
DOI https://doi.org/10.2147/CMAR.S222085
Checked for plagiarism Yes
Review by Single anonymous peer review
Peer reviewer comments 2
Editor who approved publication: Dr Sanjeev K. Srivastava
Xian Zou,1,* Zhi Heng Guo,2,* Qun Li,3 Pei Song Wang3
1NHC Key Laboratory of Nuclear Medicine, Jiangsu Key Laboratory of Molecular Nuclear Medicine, Department of Surgery, Jiang Yuan Hospital Affiliated to Jiangsu Institute of Nuclear Medicine, Wuxi 214063, Jiangsu, People’s Republic of China; 2Department of Obstetrics, The First Hospital of Jilin University, Changchun 130021, Jilin, People’s Republic of China; 3Department of Thyroid Surgery, The First Hospital of Jilin University, Changchun 130021, Jilin, People’s Republic of China
*These authors contributed equally to this work
Correspondence: Pei Song Wang
Department of Thyroid Surgery, The First Hospital of Jilin University, No. 71 on Xinmin Street, Changchun 130021, Jilin, People’s Republic of China
Tel +86 431-8187 5291
Email [email protected]
Background: Increasing evidence shows that Long non-coding RNAs (lncRNAs) involve in the development and progression processes of various cancers, including papillary thyroid cancer (PTC). In this study, we focused on the regulation function of lncRNA LINC00460 in the development of PTC.
Methods: Expression of LINC00460 was detected using quantitative real-time PCR (qRT-PCR) and Western blot assay. Cell proliferation, cell apoptosis and cell invasion were determined through CCK-8 assay, flow cytometry, and Transwell assay, respectively. In addition, target sites were detected by the dual-luciferase reporter gene assay.
Results: LINC00460 expression was markedly up-regulated in PTC tissues and cells compared to their corresponding controls by quantitative real-time PCR (qRT-PCR). Meanwhile, LINC00460 knockdown notably inhibited the proliferation capacity, accelerated the apoptosis and down-regulated the invasion-related proteins (MMP-2, MMP-9, ZEB1) expression. In addition, bioinformatics tools predicted that miR-539 both targeted with the 3ʹ-UTR of LINC00460 and MMP-9, which was confirmed by luciferase reporter assay and Western blot.
Conclusion: These findings indicated that LINC00460 can modulate MMP-9 expression to promote cell proliferation, invasion and apoptosis through targeting miR-539, suggesting act as an oncogenic RNA in PTC and provide a new therapeutic perspective.
Keywords: papillary thyroid cancer, LINC00460, proliferation, invasion, apoptosis
Introduction
TC accounts for only 1% of all the malignancies and is a common endocrine malignancy.1 Papillary thyroid carcinoma (PTC) accounts for 86% of all TC.2 Moreover, TC of different pathological patterns, the mechanism of disease occurrence is also different from each other.3 Thus, it is important to study the underlying mechanisms of PTC carcinogenesis, and it is indispensable for the development of more effective diagnostic and therapeutic methods in the future.
Recently, researchers have attempted to explore lncRNAs as a possible regulator in tumorigenesis and tumor development. The structural composition of lncRNAs is a series of non-coding RNAs >200 nucleotides in length. Previous literature has shown that lnc RNAs play a crucial regulator of tumor suppression or carcinogenesis under different circumstances. For instance, lncRNA MALAT1, lncRNA HOTAIR, lncRNA 00460, lncRNA TUG1 and lncRNA XIST, promote cell proliferation, invasion and/or migration, including multiple myeloma, triple-negative breast cancer, cervical cancer, pancreatic cancer, bladder cancer, gastric cancer and thyroid cancer.4–12
Interestingly, lncRNA can play a regulatory role in cancer development by interacting with miRNAs. MiRNAs are a series of non-coding RNAs containing about 22 nucleotides that act as competitive endogenous RNAs (ceRNAs).13,14 LINC00460 is a new cancer-associated lncRNA whose expression is involved in the development of a variety of human malignancies, including nasopharyngeal carcinoma, lung cancer, thyroid cancer.15–17 Unfortunately, the specific mechanism of action of LINC00460 in PTC is still not clear.
The purpose of this study was to investigate the biological role of LINC00460 in PTC and to determine its underlying mechanisms. LINC00460 is significantly up-regulated in PTC tissues and cell lines in comparison to their corresponding controls. LINC00460 knockdown repressed PTC cell proliferation, invasion and apoptosis. Additionally, our data revealed that LINC00460 could function as an oncogene through up-regulating the miR-539 targeted gene MMP-9 by functioning as a competitive endogenous RNA (ceRNA) for miR-539 in PTC. Our results suggest that LINC00460 may be a new indicator for diagnosis and treatment of PTC patients.
Materials and Methods
Ethical Statement and Clinical Specimens
This study was approved by the Ethical Committee of The First Hospital of Jilin University (Changchun, China). Written informed consent forms were obtained from all patients, in accordance with the Declaration of Helsinki.
Cell Culture and Transfection
Human TC cell lines (K1, TPC-1) and human normal thyroid cell line (Nthy-ori 3–1) (Shanghai Tiancheng Technology Co., Ltd.) were used in our study. The cell lines were maintained in DMEM culture medium (Hyclone, Thermo, San Jose, CA), supplemented with 10% fetal bovine serum (FBS, Gibco, USA), 100U/mL penicillin and 100μg/mL streptomycin (Invitrogen, Carlsbad, CA, USA), and incubated in a humidified chamber supplemented with 5% CO2 at 37°C.
siRNA targeting LINC00460, miR-539 mimics and miR-539 inhibitor (Life Technologies, Carlsbad, CA, USA) were, respectively, transfected using Lipofectamine 3000 Reagents (Life Technologies, Carlsbad, CA, USA). The sequences were as follows: si-LINC00460-1: 5ʹ-GUGUCAACAACCUGUUUAAUU-3ʹ; si-LINC00460- 2: 5ʹ-UUAAGUUCAGAAUUGGCACUU-3ʹ; si-LINC00460-3: 5ʹ-GUAACAACUUC UAGAGCUUUU-3ʹ. miR-539 mimics, forward, 5ʹ-UGGCAGUGUCUUAGCUGGU UG-3ʹ; reverse, 5ʹ-AC CAGCUAAGACACUGCCAUU-3ʹ. miR-539 inhibitor, 5ʹ-AC AUGG UUAGAUCA AGCACAA-3ʹ.
Quantitative RT-PCR (qRT-PCR)
Total RNA was extracted from tissues and cells using Trizol reagent (Life Technologies, Carlsbad, CA, USA). Then, RNA was reverse transcribed into cDNA. Finally, qRT-PCR experiments were conducted using SYBR Premix Ex Taq (Qiagen, Hilden, Germany). All primers were presented as follows: LINC00460, forward: 5ʹ-A ATGGTGGTAGGAGGGAGGA-3ʹ, reverse:5ʹ-CAAGGGGAATGAACACGAGG-3ʹ; β-actin, forward: 5ʹ-AAGCCCACTTCTCTCCACCTAA-3ʹ, reverse: 5ʹ-AATGCTAT CACCTCCCCTGTGT-3ʹ.
Cell Proliferation Assay
PTC cell lines (K1, TPC-1) (2x103cells/well) were transfected with siRNAs, and subsequently seeded in 96-well cell culture plates with five replicate wells for each group for 72 h. 10 μL of CCK-8 reagent was added into each well for 2 h and absorbance was measured at 450nm.
Cell Apoptosis Assay
Cell apoptosis was determined utilizing Annexin V-fluorescein isothiocyanate (FITC)/propidium iodide (PI) staining (BD Bioscience, San Jose, CA, USA). Briefly, the PTC cell lines (K1, TPC-1) (2x106cells/mL) were harvested in 6-well plates and suspended in binding buffer for 15 min containing 5 µL of Annexin V-FITC and PI. Stained cells were measured by a flow cytometer (BD LSRII; BD Pharmingen).
Cell Invasion Assay
Transwell chambers with 8 mm pores were precoated with Matrigel matrix (BD Bioscience, Franklin Lakes, NJ, USA). Transfected cells (K1, TPC-1) (1x104) were seeded into the upper chambers containing serum-free DMEM, while the lower chambers were filled with medium containing 10% FBS as a chemoattractant. After incubation for 24 h, the non-invasive cells on the top side of the membrane were removed and the invasive cells of the inserts on the lower surface were fixed and stained with 0.1% crystal violet (Sigma, St Louis, MO, USA). The number of cells was counted in six fields under a light microscope.
Western Blotting
Protein extracts from PTC cells were electrophoresed using 10% SDS-PAGE and transferred to PVDF membranes (Millipore, Billerica, MA, USA). Then, the membrane was blocked with 5% skimmed milk for 1 h followed by incubation with the following primary antibodies at 4°C overnight: Rabbit anti-MMP-2 (1:800, ab37150, Abcam, Cambridge, England, US), rabbit anti-MMP-9 (1:800, ab38898, Abcam), rabbit anti-ZEB1 (1:800, ab124512, Abcam) and Rabbit anti-β-actin (1:2000, ab37168, Abcam, Cambridge, MA, USA). Afterward, the HRP-conjugated goat anti-rabbit antibody (1:1500, Abcam) for 2 h at room temperature was added to probe the target proteins. Finally, Immunoblot signals were visualized using ECL reagents (Syngene, Frederick, MD, USA). Relative protein expression was quantified using Image-Pro Plus 6.0 software.
Luciferase Activity Assay
Wild-type (WT) or mutant controls (Mut) of 3ʹ-UTRs of the LINC00460 and MMP-9 that contained the putative miR-539 binding site were inserted into the pGL3 vector (Promega, Madison, WI, USA) for luciferase reporter experiments, named as LINC00460 WT, MMP9 WT, LINC00460 Mut and MMP-9 MMP-9 Mut, and then co-transfected into the cells with either WT or Mut, and miR-539 or control miRNA using Lipofectamine 2000 reagent for 48 h. Finally, the dual-luciferase reporter system (Promega) was used to analyze the relative luciferase activity.
Data Analysis
Data analysis was performed using GraphPad Prim 5.0 (GraphPad Software,La Jolla, CA, USA). All data are expressed as the means±standard deviation (SD). Comparisons between two groups were performed by using Student’s t-tests. P<0.05 was considered statistically significant.
Results
Expression Levels of lncRNA LINC00460 in PTC Tissues and Cells
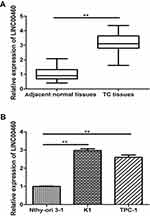
The expression levels of LINC00460 in PTC tissues and adjacent normal tissues were measured using qRT-PCR. Our data revealed that the LINC00460 expression level in PTC tissues was markedly increased than that of the adjacent normal tissues (P<0.01; Figure 1A). At the same time, LINC00460 expression levels were also obviously higher in PTC cell lines (K1, TPC-1) than that of human normal thyroid cell line Nthy-ori 3–1 (P<0.01; Figure 1B). Therefore, these data revealed that LINC00460 was markedly up-regulated in PTC tissues and cells, suggesting LINC00460 may be used as potential oncogenic role in PTC tissues and cells.
lncRNA LINC00460 Knockdown Restrained Proliferation of PTC Cells in vitro
To investigate the role of LINC00460 in the PTC process in vitro, siRNA targeting LINC00460 was transfected into PTC cell lines (K1, TPC-1) to knockdown LINC00460 expression (P<0.05; Figure 2A and B). LINC00460 knockdown obviously inhibited the proliferation of PTC cells (K1, TPC-1) using the CCK-8 assay (P<0.05; Figure 2C and D). In a word, our results found that LINC00460 knockdown inhibited the proliferation of PTC cells in vitro.
lncRNA LINC00460 Knockdown Increased Apoptosis of PTC Cells
Through flow cytometry analysis, our data revealed that LINC00460 knockdown notably increased the apoptotic rate of PTC cell lines (K1, TPC-1) compared to the control group (P<0.01; Figure 2E and F). In a word, our results indicated that LINC00460 knockdown obviously facilitated the apoptosis of PTC cells.
lncRNA LINC00460 Knockdown Restrained Invasion of PTC Cells
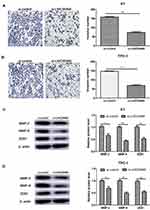
Our results revealed that LINC00460 knockdown obviously decreased the invasion number of PTC cells (K1, TPC-1) compared to the control group (P<0.01; Figure 3A and B). Moreover, LINC00460 knockdown transfection markedly down-regulated these invasion-related marker proteins (MMP-2, MMP-9, ZEB1) expression compared to control group using Western blotting (P<0.05; Figure 3C and D). In a word, our results demonstrated that LINC00460 knockdown obviously inhibited the invasion capacity of PTC cells.
lncRNA LINC00460 Enhanced MMP-9 Protein Expression via Targeting miR-539
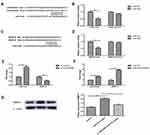
To investigate the role of LINC00460 and its downstream regulatory pathway in PTC progression, we use the bioinformatics tool to find potential target sites of LINC00460. Our results revealed that LINC00460 harbored miR-539 with complementary binding at 3ʹ-UTR regions (Figure 4A). Meanwhile, luciferase reporter revealed the luciferase activity was notably decreased (P<0.05; Figure 4B), indicating that LINC00460 could combine with miR-539. Meanwhile, it has confirmed that miR-539 targeted with the 3ʹ-UTR of MMP-9 (Figure 4C and D). After transfected with si-LINC00460 or si-NC in K1 cells, miR-539 expression was obviously increased by qRT-PCR (P<0.05; Figure 4E). However, the mRNA expression level of MMP-9 was significantly down-regulated in the LINC00460 knockdown group (P<0.05; Figure 4E). Subsequently, transfected with miR-539 inhibitor or control in K1 cells, the expression level of LINC00460 and MMP-9 was obviously up-regulated (P<0.05; Figure 4F). Western blot results revealed that miR-539 inhibitor obviously up-regulated the protein expression level of MMP-9, while the co-transfection of miR-539 inhibitor and si-LINC00460 reversed its expression levels (P<0.05; Figure 4G). In a word, these results demonstrated that LINC00460 markedly enhanced MMP-9 protein expression via targeting miR-539, suggesting that it may be achieved through the LINC00460/miR-539/MMP-9 pathway.
Discussion
Over the past decades, the aberrant expression of lncRNA has been widely observed and reported to be involved in tumorigenesis. lncRNAs have captured increasing attention due to their crucial roles in biological processes, both normal physiological processes and pathogenic processes.18–20 As an important means of epigenetic regulation, lncRNAs have been confirmed to involve in a series of human diseases, including tumors, cardiovascular and endocrine system disease.21–23 Therefore, considering the crucial role of lncRNAs in the tumorigenesis, we try to explore the effect of lncRNAs in the TC progression.
In this study, our data revealed that LINC00460 was markedly increased in PTC tissues and cell lines (K1 and TPC-1) in comparison with adjacent normal tissues and human normal thyroid cell line Nthy-ori 3–1, suggesting LINC00460 was considered as a potential oncogenic in PTC. For example, in non-small cell lung cancer (NSCLC), LINC00460 expression was obviously up-regulated and associated with poor prognosis.24 Meanwhile, previous research also confirmed that LINC00460 contributed to enhancing the capability of cell proliferation, migration and invasion through inducing ETM transition in lung cancer cells.16 Furthermore, LINC00460 was positively correlated with esophageal squamous cell carcinoma (ESCC) TNM stage, lymph node metastasis, and poor prognosis, acting as a valuable prognostic biomarker for ESCC diagnosis.17 Subsequently, further experiments explored the biological function of LINC00460 in PTC progression. Our data revealed that LINC00460 knockdown obviously restrained the proliferation and strengthened the apoptosis of PTC cell lines. In addition, LINC00460 knockdown also markedly weakened the invaded cell quantity, and down-regulated the invasion-related proteins (MMP-2, MMP-9, ZEB1) expression. In a word, these findings revealed that LINC00460 functions as a potential oncogene in PTC transformation, which might bright a new insight for the disease.
Until now, the major pattern of lncRNA regulating the tumorigenesis is the competing endogenous RNA (ceRNA).25 lncRNA functions as miRNA “sponge” to absorb miRNA abundance to reduce the expression of miRNA, and release the inhibition of target protein through miRNA.26–28 For instance, Long noncoding RNA LINC00460 promotes meningioma progression and metastasis through targeting miR-539/MMP-9.29 In our study, we confirmed that lncRNA LINC00460 modulates MMP-9 to promote cell proliferation, invasion and apoptosis by targeting miR-539 in PTC. Meanwhile, LINC00460 acts as a miR-539 sponge to restore the MMP-9 protein expression. Previous studies demonstrated that miR-539 was considered as a tumor suppressor in cancers, including breast cancer, non-small cell lung cancer, hepatocellular carcinoma.30–32 In addition, we speculate that the invasion or metastasis may more likely occur in the high grades of PTC.
Conclusion
In summary, these findings demonstrated that LINC00460 is obviously increased in PTC tissue and cells. LINC00460 enhances MMP-9 expression by targeting miR-539 to facilitate the proliferation and metastasis of PTC, providing a new treatment insight for the pathogenesis.
Abbreviations
LINC00460, long intergenic noncoding RNA-00460; PTC, papillary thyroid cancer; MMP-9, matrix metalloprotein-9; miR-539, microRNA-539; FBS, fetal bovine serum.
Acknowledgment
The study was supported by Natural Science Foundation of Jilin Province (Grant No:20180101138JC) and Wuxi Municipal Science and Technology Development Planning Funds (CSE31N1728).
Disclosure
All authors declare no conflicts of interest in this study.
References
1. Xing M. Molecular pathogenesis and mechanisms of thyroid cancer. Nat Rev Cancer. 2013;13(3):184–199. doi:10.1038/nrc3431
2. Yang L, Zheng R, Wang N, Zhang S, Chen W. [Analysis of incidence and mortality of thyroid cancer in China, 2010]. Zhonghua Yu Fang Yi Xue Za Zhi. 2014;48(8):663–668.
3. Zhang X, Mao H, Lv Z. microRNA role in thyroid cancer pathogenesis. Front Biosci (Landmark Ed). 2013;18:734–739. doi:10.2741/4135
4. Gu Y, Xiao X, Yang S. LncRNA MALAT1 acts as an oncogene in multiple myeloma through sponging miR-509-5p to modulate FOXP1 expression. Oncotarget. 2017;8(60):101984–101993. doi:10.18632/oncotarget.21957
5. Zuo Y, Li Y, Zhou Z, Ma M, Fu K. Long non-coding RNA MALAT1 promotes proliferation and invasion via targeting miR-129-5p in triple-negative breast cancer. Biomed Pharmacother. 2017;95:922–928. doi:10.1016/j.biopha.2017.09.005
6. Wu X, Cao X, Chen F. LncRNA-HOTAIR activates tumor cell proliferation and migration by suppressing miR-326 in cervical cancer. Oncol Res. 2017. doi:10.3727/096504017X15037515496840
7. Qin CF, Zhao FL. Long non-coding RNA TUG1 can promote proliferation and migration of pancreatic cancer via EMT pathway. Eur Rev Med Pharmacol Sci. 2017;21(10):2377–2384.
8. Hu Y, Deng C, Zhang H, Zhang J, Peng B, Hu C. Long non-coding RNA XIST promotes cell growth and metastasis through regulating miR-139-5p mediated Wnt/beta-catenin signaling pathway in bladder cancer. Oncotarget. 2017;8(55):94554–94568. doi:10.18632/oncotarget.21791
9. Liu H, Deng HY, Zhao YJ, Li C, Liang Y. LncRNA XIST/miR-34a axis modulates the cell proliferation and tumor growth of thyroid cancer through MET-PI3K-AKT signaling. J Exp Clin Canc Res. 2018;37(1):279. doi:10.1186/s13046-018-0950-9
10. Wang F, Liang S, Liu X, Han L, Wang J, Du Q. LINC00460 modulates KDM2A to promote cell proliferation and migration by targeting miR-342-3p in gastric cancer. Onco Targets Ther. 2018;11:6383–6394. doi:10.2147/OTT.S169307
11. Di W, Li Q, Shen W, Guo H, Zhao S. The long non-coding RNA HOTAIR promotes thyroid cancer cell growth, invasion and migration through the miR-1- CCND2 axis. Am J Cancer Res. 2017;7(6):1298–1309.
12. Lei H, Gao Y, Xu X. LncRNA TUG1 influences papillary thyroid cancer cell proliferation, migration and EMT formation through targeting miR-145. Acta Biochim Biophys Sin Shanghai. 2017;49(7):588–597. doi:10.1093/abbs/gmx047
13. Jalali S, Bhartiya D, Lalwani MK, Sivasubbu S, Scaria V. Systematic transcriptome wide analysis of lncRNA-miRNA interactions. PLoS One. 2013;8(2):e53823. doi:10.1371/journal.pone.0053823
14. Salmena L, Poliseno L, Tay Y, Kats L, Pandolfi PP. A ceRNA hypothesis: the Rosetta stone of a hidden RNA language? Cell. 2011;146(3):353–358. doi:10.1016/j.cell.2011.07.014
15. Kong YG, Cui M, Chen SM, Xu Y, Xu Y, Tao ZZ. LncRNA-LINC00460 facilitates nasopharyngeal carcinoma tumorigenesis through sponging miR-149-5p to up-regulate IL6. Gene. 2018;639:77–84. doi:10.1016/j.gene.2017.10.006
16. Li K, Sun D, Gou Q, et al. Long non-coding RNA linc00460 promotes epithelial-mesenchymal transition and cell migration in lung cancer cells. Cancer Lett. 2018;420:80–90. doi:10.1016/j.canlet.2018.01.060
17. Liang Y, Wu Y, Chen X, et al. A novel long noncoding RNA linc00460 up-regulated by CBP/P300 promotes carcinogenesis in esophageal squamous cell carcinoma. Biosci Rep. 2017;37:5. doi:10.1042/BSR20171019
18. Li X, Cao Y, Gong X, Li H. Long noncoding RNAs in head and neck cancer. Oncotarget. 2017;8(6):10726–10740. doi:10.18632/oncotarget.12960
19. Gutschner T, Diederichs S. The hallmarks of cancer: a long non-coding RNA point of view. RNA Biol. 2012;9(6):703–719. doi:10.4161/rna.20481
20. Crea F, Watahiki A, Quagliata L, et al. Identification of a long non-coding RNA as a novel biomarker and potential therapeutic target for metastatic prostate cancer. Oncotarget. 2014;5(3):764–774. doi:10.18632/oncotarget.1769
21. Zhang Z, Wang X, Cao S, et al. The long noncoding RNA TUG1 promotes laryngeal cancer proliferation and migration. Cell Physiol Biochem. 2018;49(6):2511–2520. doi:10.1159/000493876
22. Wu Y, Yuan T, Wang WW, et al. Long noncoding RNA HOST2 promotes epithelial-mesenchymal transition, proliferation, invasion and migration of hepatocellular carcinoma cells by activating the JAK2-STAT3 signaling pathway. Cell Physiol Biochem. 2018;51(1):301–314. doi:10.1159/000495231
23. Long B, Li N, Xu XX, et al. Long noncoding RNA FTX regulates cardiomyocyte apoptosis by targeting miR-29b-1-5p and Bcl2. Biochem Biophys Res Commun. 2018;495(1):312–318. doi:10.1016/j.bbrc.2017.11.030
24. Yue QY, Zhang Y. Effects of Linc00460 on cell migration and invasion through regulating epithelial-mesenchymal transition (EMT) in non-small cell lung cancer. Eur Rev Med Pharmacol Sci. 2018;22(4):1003–1010. doi:10.26355/eurrev_201802_14382
25. Zhang Y, Liu X, Li Q, Zhang Y. lncRNA LINC00460 promoted colorectal cancer cells metastasis via miR-939-5p sponging. Cancer Manag Res. 2019;11:1779–1789. doi:10.2147/CMAR.S192452
26. Cui C, Zhai D, Cai L, Duan Q, Xie L, Yu J. Long noncoding RNA HEIH promotes colorectal cancer tumorigenesis via counteracting miR-939-mediated transcriptional repression of Bcl-xL. Cancer Res Treat. 2018;50(3):992–1008. doi:10.4143/crt.2017.226
27. Liu S, Yan G, Zhang J, Yu L. Knockdown of long noncoding RNA (lncRNA) Metastasis-Associated Lung Adenocarcinoma Transcript 1 (MALAT1) inhibits proliferation, migration, and invasion and promotes apoptosis by targeting miR-124 in retinoblastoma. Oncol Res. 2018;26(4):581–591. doi:10.3727/096504017X14953948675403
28. Lv J, Wang L, Zhang J, et al. Long noncoding RNA H19-derived miR-675 aggravates restenosis by targeting PTEN. Biochem Biophys Res Commun. 2018;497(4):1154–1161. doi:10.1016/j.bbrc.2017.01.011
29. Xing H, Wang S, Li Q, Ma Y, Sun P. Long noncoding RNA LINC00460 targets miR-539/MMP-9 to promote meningioma progression and metastasis. Biomed Pharmacother. 2018;105:677–682. doi:10.1016/j.biopha.2018.06.005
30. Gao X, Li S, Li W, et al. MicroRNA-539 suppresses tumor cell growth by targeting the WNT8B gene in non-small cell lung cancer. J Cell Biochem. 2017;46(8):567–575. doi:10.1002/jcb.26634
31. Guo J, Gong G, Zhang B. miR-539 acts as a tumor suppressor by targeting epidermal growth factor receptor in breast cancer. J Cell Biochem. 2018;8(1):2073–2082. doi:10.1038/s41598-018-20431-z
32. Liu Y, Hong W, Zhou C, et al. miR-539 inhibits FSCN1 expression and suppresses hepatocellular carcinoma migration and invasion. Oncol Rep. 2017;37(5):2593–2602. doi:10.3892/or.2017.5549
 © 2020 The Author(s). This work is published and licensed by Dove Medical Press Limited. The full terms of this license are available at https://www.dovepress.com/terms.php and incorporate the Creative Commons Attribution - Non Commercial (unported, v3.0) License.
By accessing the work you hereby accept the Terms. Non-commercial uses of the work are permitted without any further permission from Dove Medical Press Limited, provided the work is properly attributed. For permission for commercial use of this work, please see paragraphs 4.2 and 5 of our Terms.
© 2020 The Author(s). This work is published and licensed by Dove Medical Press Limited. The full terms of this license are available at https://www.dovepress.com/terms.php and incorporate the Creative Commons Attribution - Non Commercial (unported, v3.0) License.
By accessing the work you hereby accept the Terms. Non-commercial uses of the work are permitted without any further permission from Dove Medical Press Limited, provided the work is properly attributed. For permission for commercial use of this work, please see paragraphs 4.2 and 5 of our Terms.