Back to Journals » OncoTargets and Therapy » Volume 14
Long Noncoding RNA JAKMIP2-AS1 Promotes the Growth of Colorectal Cancer and Indicates Poor Prognosis
Authors Ma T , Qiao T, Yuan Z, Wang G , Huang R, Wang M, Hu H, Zhu Y, Zou X, Wang X
Received 10 November 2020
Accepted for publication 22 January 2021
Published 3 February 2021 Volume 2021:14 Pages 763—772
DOI https://doi.org/10.2147/OTT.S289617
Checked for plagiarism Yes
Review by Single anonymous peer review
Peer reviewer comments 3
Editor who approved publication: Prof. Dr. Nicola Silvestris
Tianyi Ma,1 Tianyu Qiao,1 Ziming Yuan,1 Guiyu Wang,1 Rui Huang,1 Meng Wang,1 Hanqing Hu,1 Yihao Zhu,1 Xiaoming Zou,2 Xishan Wang1,3
1Department of Colorectal Surgery, The Second Affiliated Hospital of Harbin Medical University, Harbin, 150001, People’s Republic of China; 2Department of General Surgery, The Second Affiliated Hospital of Harbin Medical University, Harbin, 150001, People’s Republic of China; 3Department of Colorectal Surgery, Cancer Institute and Hospital, Chinese Academy of Medical Sciences, Peking Union Medical College, Beijing, 100021, People’s Republic of China
Correspondence: Xiaoming Zou; Xishan Wang Email [email protected]; [email protected]
Background: The identification of cancer-associated long noncoding RNAs and the investigation of their molecular and biological functions are important for understanding the molecular biology and progression of cancer. JAKMIP2-AS1 has not been reported in the literature, especially in the context of colorectal cancer. The aim of the present study was to examine the expression pattern of JAKMIP2-AS1 in colorectal cancer (CRC) and evaluate its biological role and clinical significance in tumor progression.
Methods: JAKMIP2-AS1 expression was analyzed in 56 CRC tissues and nine CRC cell lines by quantitative reverse-transcription polymerase chain reaction (qRT-PCR). Overexpression and RNA interference (RNAi) approaches were used to investigate the biological functions of JAKMIP2-AS1. The effect of JAKMIP2-AS1 on proliferation was evaluated by CCK-8, colony formation, and EdU assays. Subcutaneous injection of cells was used to study proliferation in BALB/c nude male mice. Proliferation-related protein levels were examined by immunohistochemical analysis. Differences between groups were tested for significance using Student’s t-test (two-tailed).
Results: JAKMIP2-AS1 was highly expressed in both CRC samples and cell lines compared with the corresponding normal counterparts. The upregulation of JAKMIP2-AS1 expression promoted the proliferation of colorectal cancer cells. Moreover, patients with high levels of JAKMIP2-AS1 expression had a relatively poor prognosis. Inhibition of JAKMIP2-AS1 by RNAi decreased the proliferation of CRC cells in vitro and impeded cell growth in vivo. Ki-67 and PCNA levels were affected by JAKMIP2-AS1 knockdown or overexpression in vivo.
Conclusion: Our findings indicate that JAKMIP2-AS1 is significantly upregulated in CRC tissues and regulates CRC cell proliferation. Thus, JAKMIP2-AS1 may represent a new marker of poor prognosis and is a potential therapeutic target for CRC intervention.
Keywords: colorectal cancer, long noncoding RNA, proliferation, prognosis
Introduction
Colorectal cancer (CRC) is a widely distributed disease worldwide, and its morbidity and mortality increase yearly.1 Thus, a detailed understanding of the mechanisms underlying CRC development and progression is essential for improving the diagnosis, prevention and treatment of this disease.2–4 Early stage colorectal cancer has a 5 year survival rate of 90%, but Stage ш and Stage Ⅳ colorectal cancer has a relatively low survival rate. Although many new treatments and surgical methods have appeared in recent years, such as immunotherapy and total mesorectal excision, there has been no major change in patient survival.5 Therefore, further research on the pathogenesis of colorectal cancer is urgently needed.
Long noncoding RNAs are a class of noncoding RNAs that are more than 200 nucleotides in length.6,7 Their functions cover many areas, including cell heterogeneity, proliferation and tumor formation.8,9 For a long time, long noncoding RNAs have been regarded as dark matter in the gene transcription process. With the deepening of research, long noncoding RNAs have been determined to play an important role in biological processes.10 Especially in the process of tumor development, long noncoding RNA is now used as a new molecular marker or therapeutic target.11
The long noncoding RNA JAKMIP2-AS1 is located in the human genome at 19q13.41 and has been identified as a noncoding RNA with 1920 nucleotides.12 There are no reports about this gene and malignant tumors. The RNA is highly expressed in the testis and placenta but less expressed in the colon, duodenum and liver.12 In the current study, we discovered for the first time that JAKMIP2-AS1 is highly expressed in colorectal cancer and is related to tumor growth and patient prognosis.
Methods
Tissue Collection
All human CRC tissues and paracancerous normal tissues were collected in the Department of Colorectal Cancer Surgery, the Second Affiliated Hospital of Harbin Medical University. Patients were pathologically and clinically diagnosed with colorectal cancer. Informed consent was obtained from the patients before sample collection in accordance with institutional guidelines. This study was carried out under permission of the Clinical Research Ethics Committees of the Second Affiliated Hospital of Harbin Medical University. The clinical characteristics are shown in Table 1.
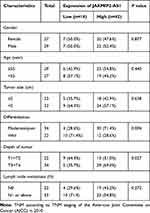 |
Table 1 Correlation Between JAKMIP2-AS1 Expression and Clinicopathological Characteristics of Colorectal Cancer Patients |
Cell Culture
NCM460 and LOVO cells were purchased from the Beijing BeiNa Chuanglian Biotechnology Research Institute (Beijing China). Human CRC cell lines (SW1116, SW480, SW620, HCT116, HT29, RKO and DLD1) and a normal colon epithelium cell line (NCM460) were purchased from the American Type Culture Collection (ATCC, Manassas, VA, USA). NCM460 human colonic normal epithelial cells and the human colon cancer cell lines HT29, LOVO, RKO and DLD1 were cultured in RPMI medium (GIBCO, USA) in a humidified incubator at 37 °C under 5% CO2 conditions, and 10% (volume/volume) fetal bovine serum (FBS) was added. Human colon cancer SW1116 cells were cultured in L15 medium (GIBCO, USA), cultured in air at 37 °C in a humidified incubator, and supplemented with 10% (volume/volume) fetal bovine serum (FBS). Human colon cancer HCT116 cells were cultured in McCoy's 5A medium (Jiangsu Kaiji Biotechnology Co., Ltd.), cultured in a humidified incubator at 37 °C under 5% CO2 conditions, and supplemented with 10% (volume/volume) fetal bovine serum (FBS). Human colon cancer SW480 and SW620 cells were cultured in DMEM high-sugar medium (GIBCO, USA) in a humidified incubator at 37 °C under 5% CO2, and 10% (volume/volume) fetal bovine serum (FBS) was added.
Construction of Stable Cell Lines
A lentivirus (Shanghai Genechem Co., Ltd., Shanghai, China) was used to construct a cell line overexpressing JAKMIP2-AS1 (Lenti-JAKMIP2-AS1) and a corresponding empty vector [Lenti-negative control (NC)]. A total of 1 day prior to lentiviral infection, DLD-1 cells were seeded in 6-well plates at 2x105 cells/well and cultured in a cell incubator containing 5% CO2 at 37 °C for 18–24 h so that the cells reached 70%~80% plate bottom area at the time of transfection. Lenti-JAKMIP2-AS1 was transfected into cells using polybrene (2 μg/mL). Using the same method for the Lenti-negative control, the corresponding Lenti-NC was introduced into cells and incubated at 37 °C in an incubator containing 5% CO2 for 4–6 h; subsequently, fresh medium was added to the cells. To establish a stable cell line, cells were selected after 96 h in fresh medium by replacing the medium with medium containing puromycin (1–2 mg/mL; Sigma-Aldrich; Merck KGaA, Darmstadt, Germany); the cells were cultured with puromycin for at least 1 week after infection, and cells were used for mRNA analyses and other tests.
Cell Transfection
We selected the HT29 cell line for the knockdown study based on the expression of JAKMIP2-AS1 in the CRC cell line. The cells were cultured in culture dishes to a semifused state, and JAKMIP2-AS1 and negative control siRNAs (GenePharma, Shanghai, China) were transfected with Lipofectamine 2000 (Invitrogen, USA) for 24 h. A confocal fluorescence microscope (OLYMPUS, JAPAN) was used to detect fluorescently labeled siRNAs and to examine the transfection efficiency. qPCR was used to verify the knockdown efficiency of JAKMIP2-AS1.
RNA Extraction and Quantitative Real-Time PCR
Total RNA was extracted from tissues and cells using TRIzol reagent (Invitrogen, USA), and DNA was quantified using a Nanodrop 2000 spectrophotometer (Thermo Fisher Scientific, Waltham, MA). Quantitative PCR (qPCR) was performed using 0.8 μg of cDNA as template with SYBR Green reagent (Thermo Fisher Scientific, Waltham, MA) using the Applied Biosystem 7500 qPCR system (Applied Biosystems). Relative expression was compared by the 2-ΔΔCt method, and β-actin served as the endogenous control. Primer sequences were as follows: JAKMIP2-AS1: forward, 5´-TTCCTGCTCGTGCTGCTTGATAAG- 3´ reverse, 5´-CACGGTGCTGGTGACAGAACTC-3´. β-actin: forward, 5´-CCTGGCACCCAGCACAAT-3´ reverse, 5´-GGGCCGGACTCGTCATAC-3´.
Cell Proliferation Assay
CCK-8 solution (Dojindo, Tokyo, Japan) was used to evaluate cell viability. After seeding 5000 transfected cells/well into 96-well plates, the proliferation of HT29 cells was assessed at three time points (24, 48, and 72 h) by incubating them with 10 µL of CCK-8 solution for 1 h at 37 °C. A microplate reader (Tecan, Switzerland) was used to quantify the absorbance of each well. The optical density (OD) was measured at 450 nm.
Colony Formation
A total of 1×103 cells were seeded in a 6-well plate (Corning, Life Sciences, USA), and then 2 mL of the corresponding medium was supplemented with 10% FBS. After 14 days, the cells were fixed at room temperature with 4% paraformaldehyde for 15 minutes, washed with PBS, and stained with 0.1% crystal violet for 15 minutes. Photos were taken after drying, and colonies were counted using ImageJ software.
EdU Assay
The treated cells were resuspended in a 24-well plate, and EdU was added to the cell culture medium to label the proliferative cells. After 2h of incubation, the cells were fixed with 4% paraformaldehyde, and glycine was added to neutralize the excess paraformaldehyde. After washing with PBS, Apollo (RiboBio, Guangzhou, China) staining reaction solution was added in the dark and incubated at room temperature for 30 minutes on a decolorization shaker. The permeate was then added to the decolorization shaker for 20 minutes. Hoechst 33342 reaction solution was added in the dark and incubated for 30 minutes on the decolorization shaker.
Tumorigenesis in Nude Mice
BALB/c nude mice (4–6 weeks) were housed and maintained under specific pathogen-free conditions and used in accordance with institutional guidelines approved by the Animal Care and Use Committee. For xenografts, 1×107 HT29 NC and siRNA-1836 cells were subcutaneously injected into nude mice (n=5 for each group), and 1×107 DLD-1 NC and lenti-JAKMIP2-AS1 cells were subcutaneously injected into nude mice (n=5 for each group). Mice were euthanized 25 days after tumor formation, and tumors were excised. The tumor volume was evaluated every 5 days using a caliper based on the formula (A×B2)/2, where A is the maximum diameter and B is the diameter perpendicular to A. After 25 days, tumor xenografts were fixed in 4% paraformaldehyde for subsequent immunohistochemical studies. This study was carried out with the approval of the Ethics Review Committee of the Second Affiliated Hospital of Harbin Medical University.
Immunohistochemical (IHC) Analysis
Paraffin-embedded xenografts were used for IHC detection as described in previous study. Briefly, tumor samples were embedded in paraffin and sectioned into 2- to 4-µm slices, and antibodies against Ki-67 and PCNA were then used for IHC staining according to the manufacturer’s instructions. The sections were then imaged using a digital microscope camera.
Statistical Analyses
All experimental assays were conducted at least three times with samples in triplicate. Data are represented as the means ± standard deviation. The two-tailed Student’s t test was used to determine the statistical significance of differences between groups, and Pearson correlation analysis was used to assess the relationship among gene sets. One-way ANOVA followed by Tukey’s post hoc test between multiple groups were applied.Statistical analysis was performed using the Statistical Program for Social Sciences 22.0 software (SPSS, CA, USA) and was visualized using GraphPad Prism 8.0 (GraphPad Software, CA, USA). A P value < 0.05 was considered significant, and P values were indicated as follows: *P < 0.05, **P < 0.01.
Results
JAKMIP2-AS1 is Apparently Upregulated in CRC
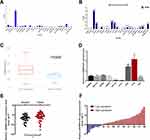
First, we browsed several online bioinformatics tools to identify lncRNAs potentially implicated in the initiation and progression of CRC. As a result, the NCBI database (https://www.ncbi.nlm.nih.gov/gene/) showed that JAKMIP2-AS levels were generally underexpressed in 26 of 27 different tissues (except brain tissues) from 95 healthy individuals, including normal colon tissues (Figure 1A). Similarly, data from the NONCODE database (http://www.noncode.org/) also showed the underexpression of JAKMIP2-AS1 in normal colon tissues (Figure 1B). However, based on the analysis of GEO data from the lnCAR website (https://lncar.renlab.org/, GEO: GSE33113, Analysis ID: CR_S131), the expression of JAKMIP2-AS1 in colorectal tumor tissues was significantly higher than that in normal tissues (Figure 1C). Based on the above relevant results, we speculate that the high expression of JAKMIP2-AS1 in colorectal cancer is involved in tumor development. Therefore, we first verified the expression of JAKMIP2-AS1 in colorectal cancer cell lines and normal colorectal cell lines. The expression of JAKMIP2-AS1 increased significantly in LOVO and HT29 cell lines, while the expression of DLD-1 and HCT116 decreased (Figure 1D). Furthermore, HT29 cells expressed the highest levels of JAKMIP2-AS1, while DLD-1 cells expressed the lowest levels among the eight CRC cell lines. These cells were selected as representative CRC cell lines for subsequent studies.
Analysis of the expression of JAKMIP2-AS1 in colorectal cancer tissues in our center showed that the expression in tumor tissues was higher than that in normal control tissues (Figure 1E), consistent with the results of the database. The patients were divided into JAKMIP2-AS1 high and JAKMIP2-AS1 low groups according to JAKMIP2-AS1 expression to investigate the significance of aberrant expression of JAKMIP2-AS1 in CRC (Figure 1F). The statistical results showed that the expression of JAKMIP2-AS1 was highly correlated with differentiation, depth of tumor and lymph node metastasis (Table 1).
JAKMIP2-AS1 Promotes Colorectal Cancer Cell Propagation
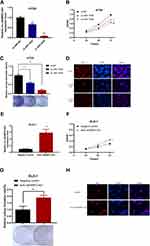
The high expression of JAKMIP2-AS1 in tissues prompted us to study the function of JAKMIP2-AS1 in tumor cell development. Subsequently, to study the function of JAKMIP2-AS1 in the development of colorectal cancer, we knocked down JAKMIP2-AS1 in HT29 cells and overexpressed it in DLD-1 cells. As a result, we can see that siRNA-1635 and siRNA-1836 have higher knockdown efficiencies (Figure 2A).
To verify the function of JAKMIP2-AS1 in colorectal cancer, we performed experiments such as CCK-8, colony formation, sphere, transwell, wound healing, and apoptosis assays to verify the role of the molecule in cell growth. However, except for the CCK-8 and colony formation assays, which showed statistical significance, no differences were detected in the other assays. The results of the CCK-8 assay showed that the growth curve of the JAKMIP2-AS1 knockdown group was different from that of the control group (Figure 2B). We obtained the same result in the cloning experiment. Knocking down JAKMIP2-AS1 affected the colony formation ability of HT29 cells (Figure 2C). To verify this proliferation capacity, we subsequently performed an EdU experiment. The results showed that compared with the control group, there was less red fluorescence in the EdU assay when JAKMIP2-AS1 was knocked down (Figure 2D).
Therefore, JAKMIP2-AS1 was stably overexpressed in DLD-1 cell lines by lentiviral transduction (Figure 2E). As expected, the growth curve of JAKMIP2-AS1 showed the same trend after overexpression (Figure 2F). After stable overexpression of JAKMIP2-AS1, the number of cells in the cell dish increased significantly compared with the control group in the colony formation assay (Figure 2G). At the same time, we also observed that the red fluorescence increased after overexpression of JAKMIP2-AS1 compared to the control group (Figure 2H). The results of this section show that the silencing of JAKMIP2-AS1 inhibits the growth of tumor cells.
JAKMIP2-AS1 Affects the Proliferation of Colorectal Cancer in vivo
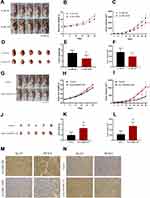
After the above experiments, we initially confirmed that JAKMIP2-AS1 affects tumor proliferation in colorectal cancer cell lines. To investigate whether the molecule affects tumor in vivo proliferation, we constructed a mouse model of JAKMIP2-AS1 knockdown. The results showed that the tumors in mice injected with treated HT29 cells were smaller than those in the control group (Figure 3A). The mice injected with HT29 cells after knocking down JAKMIP2-AS1 weighed slightly less than the control group (Figure 3B). However, in the mice in the treatment group, the size of the tumor was significantly lower than in the control group (Figure 3C). After the tumors were removed from the mice, the tumors in the siRNA-1836 group were smaller than those in the control group (Figure 3D). There were also differences in the weight and volume of the tumor (Figure 3E and F).
In the next experiment, we injected the overexpressed DLD-1 cells subcutaneously into mice to obtain photos of the control group (Figure 3G). As shown in the picture, DLD-1 mice injected with overexpressed JAKMIP2-AS1 showed greater tumor growth than control mice. Following statistical analysis of the relevant test data after tumor formation in mice, it was evident that the weight gain of the mice in the overexpression group was smaller (Figure 3H). However, the volume of the tumor in the overexpression group was significantly increased compared with that in the control group (Figure 3I). The tumors of the mice in the JAKMIP2-AS1 overexpression group were significantly larger than those of the control group (Figure 3J). There were also significant differences in the weight and volume of tumors (Figure 3K and L).
JAKMIP2-AS1 Affects the Levels of Ki-67 and PCNA
The results of immunohistochemical analysis after resection of the tumors showed that Ki67 and PCNA expression levels were significantly increased compared with the control group after overexpression of JAKMIP2-AS1 (Figure 3M and N). Based on the above results, we believe that JAKMIP2-AS1 may affect tumor proliferation in vivo.
Discussion
CRC ranks among the most common and lethal malignant diseases. The poor prognosis of early-stage CRC is crucially linked to the onset of tumor proliferation.13–15 The processes inducing and stimulating accumulation are complex and still not well understood. Bian et al16 established a role for lncRNAs in the proliferation pattern in CRC. They identified lncRNA-FEZF1-AS1 as a prognostic marker for proliferation and patient survival in CRC. Similarly, Xu, M et al17 also elucidated the effects of lncRNA-SNHG1 in the proliferative ability of colorectal cancer. However, the roles of lncRNA JAKMIP2-AS1 in CRC have not been elucidated.
In this study, we observed that JAKMIP2-AS1 was highly expressed in colorectal cancer tissues from patients and was also associated with poor prognosis. Thus, JAKMIP2-AS1 may be related to the occurrence and development of colorectal cancer. We found differences in JAKMIP2-AS1 expression in normal and colorectal cancer tissues by querying relevant databases. Subsequently, we confirmed the difference in expression levels through qPCR experiments of samples from our center. Among 56 pairs of samples, 75% of cancer tissues had higher expression of JAKMIP2-AS1 than the paired normal tissues (Table 1). Meanwhile, a study of the GEO database found that patients with high JAKMIP2-AS1 expression had a worse prognosis. This difference in expression has greatly aroused our interest.
We hypothesized that JAKMIP2-AS1 plays a role in colorectal cancer development. After that, we detected the expression of JAMIP2-AS1 in colorectal cancer cell lines. We selected the HT29 cell line with higher JAKMIP2-AS1 expression and the DLD-1 cell line with lower expression for subsequent experiments. In the HT29 cell line, we knocked down JAKMIP2-AS1 by transient transfection, and in the DLD-1 cell line, we used a stable overexpression method to increase JAMIP2-AS1 expression. Knockdown of JAKMIP2-AS1 was used to investigate JAKMIP2-AS1 biological functions. Unfortunately, in the apoptosis experiment, the results obtained in the cell invasion experiment were not statistically significant. After knocking down JAKMIP2-AS1, the proliferation ability of colorectal tumor cells decreased. In contrast, the experiment showed the opposite result after overexpression of JAKMIP2-AS1.
Since JAKMIP2-AS1 has not been reported in the literature, to confirm the function of JAKMIP2-AS1 in promoting the proliferation of colorectal cancer cells, we carried out colony formation experiments. Similar to CCK-8, the number of cells in the cell dish decreased significantly after knocking down JAKMIP2-AS1. After JAKMIP2-AS1 was overexpressed, the number of cells in dishes increased significantly. Researchers often use EdU experiments to observe the proliferation ability of cells.18,19 Through EdU experiments, we confirmed that the proliferation ability of colorectal cancer cells was significantly reduced after JAKMIP2-AS1 knockdown. The number of fluorescent proliferating cells overexpressing JAKMIP2-AS1 was significantly increased.In this study, we performed apoptosis experiments, but no positive results were obtained. We speculate that JAKMIP2-AS1 does not affect the apoptosis of colorectal cancer, but only affects the proliferation-related signaling pathway of colorectal cancer.
It has been confirmed that the silencing or overexpression of JAKMIP2-AS1 significantly affects the proliferation of colorectal cancer cells in vitro. To further confirm the influence of JAMIP2-AS1 on the proliferation direction of colorectal cancer, we conducted in vivo experiments. We subcutaneously injected treated cells into mice to observe the size and proliferation rate of tumors. As shown in the previous results, the tumors of knockdown-treated cell lines were significantly reduced compared with the control group after injection in vivo, and the observed effect was more obvious after removal of the tumors. As predicted, after subcutaneous injection of the overexpression-treated cell line, the tumor was enlarged compared with the control group. Subsequently, we performed immunohistochemical staining of the tumor, and the results showed that Ki-67 and PCNA expression levels were significantly reduced after JAKMIP2-AS1 knockdown, while the expression increased in tumors overexpressing JAKMIP2-AS1.
To date, relevant results have confirmed that JAKMIP2-AS1 plays an important role in colorectal cancer tumor proliferation. We speculate that JAKMIP2-AS1 may change the expression levels of key molecules in colorectal cancer proliferation-related pathways or affect them through the ceRNA mechanism. In turn, the expression of related genes affects the proliferation of colorectal cancer.20–23 However, further experiments are needed to clarify the mechanism by which JAKMIP2-AS1 affects the proliferation of colorectal cancer. I hope that subsequent work can elaborate on the mechanism of JAKMIP2-AS1 in colorectal cancer, and hope that JAKMIP2-AS1 can be used as a new marker for the prognosis and diagnosis of colorectal cancer.
Conclusions
In summary, JAKMIP2-AS1 upregulation may be a negative prognostic factor for CRC patients, indicative of poor survival rates. The current study showed that JAKMIP2-AS1 may regulate the proliferation ability of CRC cells. These findings further the understanding of CRC pathogenesis and development and facilitate the development of lncRNA-directed diagnostics and therapeutics against this deadly disease.
Disclosure
The authors report no conflicts of interest in this work.
References
1. Siegel RL, Miller KD, Jemal A. Cancer statistics, 2020. CA Cancer J Clin. 2020;70(1):7–30. doi:10.3322/caac.21590
2. Kadosh ES-AI, Venkatachalam A, Venkatachalam A, et al. The gut microbiome switches mutant p53 from tumour-suppressive to oncogenic. Nature. 2020;586(7827):133–138. doi:10.1038/s41586-020-2541-0
3. Woolston AKK, Spain G, Spain G, et al. Genomic and Transcriptomic Determinants of Therapy Resistance and Immune Landscape Evolution during Anti-EGFR Treatment in Colorectal Cancer. Cancer Cell. 2019;36(1):35–50.e9. doi:10.1016/j.ccell.2019.05.013
4. Y ZX L, Liu Q, Liu Q, et al. lncRNA MIR100HG-derived miR-100 and miR-125b mediate cetuximab resistance via Wnt/β-catenin signaling. Nat Med. 2017;23(11):1331–1341. doi:10.1038/nm.4424
5. Siegel RL, Miller KD, Goding Sauer A, et al. Colorectal cancer statistics, 2020. CA Cancer J Clin. 2020;70(3):145–164. doi:10.3322/caac.21601
6. P SV B, Royce TE, Rozowsky JS, et al. Global identification of human transcribed sequences with genome tiling arrays. Science. 2004;306(5705):2242–2246. doi:10.1126/science.1103388
7. Birney ESJ, Dutta A, Guigo R, et al. Identification and analysis of functional elements in 1% of the human genome by the ENCODE pilot project. Nature. 2007;447(7146):799–816.
8. Kopp F, Mendell JT. Functional Classification and Experimental Dissection of Long Noncoding RNAs. Cell. 2018;172(3):393–407. doi:10.1016/j.cell.2018.01.011
9. Ransohoff JD, Wei Y, Khavari PA. The functions and unique features of long intergenic non-coding RNA. Nat Rev Mol Cell Biol. 2018;19(3):143–157. doi:10.1038/nrm.2017.104
10. Jarroux J, Morillon A, Pinskaya M. History, Discovery, and Classification of lncRNAs. Adv Exp Med Biol. 2017;1008:1–46. doi:10.1007/978-981-10-5203-3_1
11. Matsui M, Corey DR. Non-coding RNAs as drug targets. Nat Rev Drug Discov. 2017;16(3):167–179. doi:10.1038/nrd.2016.117
12. NCBI. https://www.ncbi.nlm.nih.gov/gene/?term=JAKMIP2-AS1.
13. Z DS Z, Yin K, Yin K, et al. Knockdown long noncoding RNA nuclear paraspeckle assembly transcript 1 suppresses colorectal cancer through modulating miR-193a-3p/KRAS. Cancer Med. 2019;8(1):261–275. doi:10.1002/cam4.1798
14. H LX W, Wang JR, Wang J-R, et al. TRAF6 inhibits colorectal cancer metastasis through regulating selective autophagic CTNNB1/β-catenin degradation and is targeted for GSK3B/GSK3β-mediated phosphorylation and degradation. Autophagy. 2019;15(9):1506–1522. doi:10.1080/15548627.2019.1586250
15. J WF H, Lan Y, Lan Y, et al. KIFC1 regulated by miR-532-3p promotes epithelial-to-mesenchymal transition and metastasis of hepatocellular carcinoma via gankyrin/AKT signaling. Oncogene. 2019;38(3):406–420. doi:10.1038/s41388-018-0440-8
16. Bian Z, Zhang J, Li M, et al. LncRNA-FEZF1-AS1 Promotes Tumor Proliferation and Metastasis in Colorectal Cancer by Regulating PKM2 Signaling. Clin Cancer Res. 2018;24(19):4808–4819. doi:10.1158/1078-0432.Ccr-17-2967
17. Xu M, Chen X, Lin K, et al. The long noncoding RNA SNHG1 regulates colorectal cancer cell growth through interactions with EZH2 and miR-154-5p. Mol Cancer. 2018;17(1):141. doi:10.1186/s12943-018-0894-x
18. JH LX Y, Wang TT, Wang -T-T, et al. The MBNL3 splicing factor promotes hepatocellular carcinoma by increasing PXN expression through the alternative splicing of lncRNA-PXN-AS1. Nat Cell Biol. 2017;19(7):820–832. doi:10.1038/ncb3538
19. Chen FCJ, Yang L, Yang L, et al. Extracellular vesicle-packaged HIF-1α-stabilizing lncRNA from tumour-associated macrophages regulates aerobic glycolysis of breast cancer cells. Nat Cell Biol. 2019;21(4):498–510. doi:10.1038/s41556-019-0299-0
20. W ZG H, Jiang N, Jiang N, et al. Long noncoding RNA BLACAT2 promotes bladder cancer-associated lymphangiogenesis and lymphatic metastasis. J Clin Invest. 2018;128(2):861–875. doi:10.1172/JCI96218
21. C WY L, Wang X, Wang X, et al. DNA-methylation-mediated activating of lncRNA SNHG12 promotes temozolomide resistance in glioblastoma. Mol Cancer. 2020;19(1):28. doi:10.1186/s12943-020-1137-5
22. W ZZ L, Liu X, Liu X, et al. The FOXN3-NEAT1-SIN3A repressor complex promotes progression of hormonally responsive breast cancer. J Clin Invest. 2017;127(9):3421–3440. doi:10.1172/JCI94233
23. Tian TLX, Pan G, Pan G, et al. Long Noncoding RNA MPRL Promotes Mitochondrial Fission and Cisplatin Chemosensitivity via Disruption of Pre-miRNA Processing. Clin Cancer Res. 2019;25(12):3673–3688. doi:10.1158/1078-0432.CCR-18-2739
 © 2021 The Author(s). This work is published and licensed by Dove Medical Press Limited. The full terms of this license are available at https://www.dovepress.com/terms.php and incorporate the Creative Commons Attribution - Non Commercial (unported, v3.0) License.
By accessing the work you hereby accept the Terms. Non-commercial uses of the work are permitted without any further permission from Dove Medical Press Limited, provided the work is properly attributed. For permission for commercial use of this work, please see paragraphs 4.2 and 5 of our Terms.
© 2021 The Author(s). This work is published and licensed by Dove Medical Press Limited. The full terms of this license are available at https://www.dovepress.com/terms.php and incorporate the Creative Commons Attribution - Non Commercial (unported, v3.0) License.
By accessing the work you hereby accept the Terms. Non-commercial uses of the work are permitted without any further permission from Dove Medical Press Limited, provided the work is properly attributed. For permission for commercial use of this work, please see paragraphs 4.2 and 5 of our Terms.