Back to Journals » Journal of Pain Research » Volume 13
Long Noncoding RNA H19 Induces Neuropathic Pain by Upregulating Cyclin-Dependent Kinase 5-Mediated Phosphorylation of cAMP Response Element Binding Protein
Authors Li K, Jiao Y, Ren X, You D, Cao R
Received 27 November 2019
Accepted for publication 17 July 2020
Published 20 August 2020 Volume 2020:13 Pages 2113—2124
DOI https://doi.org/10.2147/JPR.S240273
Checked for plagiarism Yes
Review by Single anonymous peer review
Peer reviewer comments 2
Editor who approved publication: Dr Michael Schatman
This paper has been retracted.
Kai Li, 1 Yuan Jiao, 1 Xuli Ren, 1 Di You, 1 Rangjuan Cao 2
1Department of Anesthesiology, China-Japan Union Hospital of Jilin University, Changchun 130021, Jilin, People’s Republic of China; 2Department of Hand-Surgery, China-Japan Union Hospital of Jilin University, Changchun 130021, Jilin, People’s Republic of China
Correspondence: Rangjuan Cao Department of Hand-Surgery
China-Japan Union Hospital of Jilin University, 126th Xiantai Avenue, Changchun 130021, Jilin, People’s Republic of China
Tel/ Fax +86-431-89876551
Email [email protected]
Objective: Neuropathic pain (NP) is a debilitating condition caused by nervous system injury and chronic diseases. LncRNA H19 is upregulated in many human diseases, including NP. Cyclin-dependent kinase 5 (CDK5) aggressively worsens inflammatory action and nerve damage to cause severe NP. Phosphorylated cAMP response element binding protein (CREB) is detrimental to nerves and promotes NP progression. Herein, aim of our study was to assess the mechanism of lncRNA H19.
Methods: The NP rat model was established using chronic constriction injury (CCI). Paw withdrawal threshold (PWT) tests and paw withdrawal latency (PWL) tests were performed. Then, small interfering (si)RNA against H19 was intrathecally injected into rats to suppress H19 expression. Schwann cells were isolated from NP rats and transfected with siRNA-H19 or a lentivirus (LV)-based vector expressing H19. Inflammatory factors and glial fibrillary acidic protein (GFAP) were detected. Western blot analysis was conducted to detect CDK5/p35 and p-CREB expression. Finally, H19, CDK5 and CREB phosphorylation were tested with the combination of the CDK5 inhibitor roscovitine and transfection of LV-H19 and siRNA-H19. Finally, we investigated the binding relationships between H19 and miR-196a-5p and between miR-196a-5p and CDK5 and detected the mRNA expression of miR-196a-5p and CDK5 in rats with H19 knockdown and in Schwann cells with H19 knockdown.
Results: Highly expressed H19, CDK5/p-35 and p-CREB were observed in NP rats, accompanied by obviously decreased PWT and PWL, upregulated inflammatory factors and GFAP levels, and reduced 5-HT 2A and GABAB2 expression. siRNA-H19 restored NP-related indexes and downregulated CDK5/p35 and p-CREB phosphorylation. siRNA-H19, together with the CDK5 inhibitor roscovitine, reduced CDK5 and p-CREB expression in Schwann cells isolated from NP rats. Binding sites between H19 and miR-196a-5p and between miR-196a-5p and CDK5 were identified. Silencing H19 upregulated miR-196a-5p expression and downregulated CDK5 levels.
Conclusion: Our study demonstrated that silencing H19 inhibited NP by suppressing CDK5/p35 and p-CREB phosphorylation via the miR-196a-5p/CDK5 axis, which may provide new insight into NP treatment.
Keywords: neuropathic pain, long noncoding RNA H19, cyclin-dependent kinases 5, cAMP response element binding protein phosphorylation, roscovitine
Introduction
As defined by the International Association for the Study of Pain, neuropathic pain (NP) directly results from a lesion or disease afflicting the somatosensory system, which is mostly caused by central nervous system injury and affects almost 8% of the world population.1–3 NP can be triggered when substantial alterations in damaged neurons and strongly downregulated pathways in the central nervous system occur in injured nerves.4 Clinical findings, including elaborate case history and careful examination of the features of NP, are essential for NP diagnosis; on the other hand, electrophysiological examinations, imaging techniques and punch skin biopsy are also conducive to clinical NP diagnosis.5 It is accepted that the main therapies, such as antidepressants, opioid analgesics, topical agents and anticonvulsants, and interventional treatments, including nerve blocks, spinal cord stimulation and steroid and anesthetic injection, are all beneficial options for relieving NP.6 Clarifying the individual differences in NP, whose heterogeneity can lead to inadequate treatment, is a challenge.7 In this context, novel therapeutic strategies for NP are urgently needed. With this in mind, we took a long noncoding RNA (lncRNA)-based approach to understand the underlying mechanism of NP development to develop novel intervention strategies.
LncRNAs play important roles in disease occurrence and development, and their associations with these diseases contribute to improved understanding of the pathogenesis, diagnosis and treatment of diseases.8 H19 overexpression predicts a negative prognosis in many cancers.9 H19 expression is increased in Schwann cells of the NP model after spinal nerve ligation.10 Typically, CDKs that are activated together with a regulatory subunit tend to be proline-guided serine protein kinases.11 However, CDK5 mainly functions in the central nervous system instead of the cell cycle.12 In a recent study, CDK5 is shown to be an active contributor to inducing and maintaining chronic pain from nerve damage and peripheral inflammation.13 CDK5 mediates cytoskeleton dynamics in the nervous system and is a component of pain signaling.14,15 In addition, as an endoplasmic reticulum-targeted transcription factor, CREB is a major factor in secretion regulation, metabolism and inflammation.16 CREB modulates different cellular responses whose phosphorylation serves as a major contributor to the nervous system by regulating protection, plasticity and development.17 From the above, it is reasonable to assume that there may be interactions between H19, CDK5 and CREB phosphorylation in NP progression. Thus, we conducted a series of experiments to verify this hypothesis.
Materials and Methods
Ethics Statement
This study was approved and supervised by the ethics committee of China Japan Union Hospital of Jilin University (Approval No. 2016-nsfc001). This study followed the guidelines for the care and use of laboratory animals (NIH Pub. No. 85–23, revised 1996) compiled by the National Institutes of Health. Every procedure in this experiment was approved by the laboratory animal ethics committee.
Model Establishment and Animal Grouping
Twenty-four adult Sprague-Dawley rats (220–250 g) (Animal Experiment Center, Central South Hospital of Wuhan University, Wuhan, Hubei, China, SCXK (Hubei) 2015–0025) were placed in a specific pathogen-free room with a 12-hour light-dark cycle at 21–25°C.
As previously described by Bennett and Xie, an NP rat model was established via chronic constriction injury (CCI). These rats were assigned to the CCI group, which was subjected to CCI, and the sham group, with 12 rats in each group. As the biceps femoris was bluntly dissected, one side of the sciatic nerves was exposed at the mid-thigh, and almost 6 mm of nerve away from the sciatic trifurcation was separated from the adhering tissues, and 4 loose chromic gut ligatures (4.0 gauge) were placed around it, with approximately 1 mm spacing between ligatures. The affected nerve length was approximately 4–5 mm. Rats in the sham group were treated in the same way as the CCI rats but without sciatic nerve ligation. All the biochemical indexes and tissues were obtained from the surgical side for a reliable and accurate experimental result. After the surgeries, a rising pain threshold of the NP rats suggested successful model establishment. After pain behavior tests, 6 rats from both groups were randomly selected for immunofluorescence assays, and the other 6 rats in each group were used to prepare tissue homogenates.
In the following experiment, rats were intrathecally placed with a catheter via the waist and anesthetized to maintain supine posture. To prevent cerebrospinal fluid leakage, the external orifice of the catheter was covered by a closed epidural catheter after the operation. Then rat activities after surgery were observed, and the position of the catheter was assessed. The separately housed rats were all injected with penicillin to prevent infection. Twenty-four rats with successfully intrathecally placed catheters were removed to establish NP models. On the third day after CCI operation, rats were intrathecally injected with 2 μM siRNA-H19 or siRNA-NC every two days until the 21st day after the CCI operation. On the 21st day, samples were taken according to the process mentioned above following pain behavior tests.
Pain Behavior Tests
Rats were placed on a metal mesh floor for the paw withdrawal threshold (PWT) test. Rat plantar surfaces were applied force using the von Frey filament (Bioseb, Pinellas Park, Fl, USA) 25 min later. Hind paw withdrawal indicated a positive response. The von Frey filament application lasted for 5 s, with a spacing interval of less than 15 s. In the paw withdrawal latency (PWL) test, which aimed to assess the period from stimulus to paw withdrawal, rats were placed separately on a glass plate with a heat stimulator (Bioseb) underneath to stimulate the rat plantar surfaces at most for 18 s. The PWT and PWL tests were both performed at days 3, 7, 14 and 21 after CCI operation. The naive baseline was measured on the previous day of the CCI operation.18
Dual-Luciferase Reporter Gene Assay
MicroRNA.org (http://www.microrna.org/microrna/home.do)19 and StarBase (http://starbase.sysu.edu.cn/index.php)20 analyses predicted that there were specific binding sites between H19 and miR-196a-5p and between miR-196a-5p and CDK5. The complementary binding sequence and its mutation sequence were amplified and cloned into the pmiR-GLO luciferase vector (Promega, Madison, WI, USA) to construct the wild-type (WT) plasmids H19-WT and CDK5-WT and the corresponding mutant (MUT) plasmids H19-MUT and CDK5-MUT. Using Lipofectamine 2000 (Invitrogen, Carlsbad, USA), according to the instructions, the constructed plasmids were cotransfected with miR-NC mimic or miR-196a-5p mimic into HEK293T cells (Shanghai Institute of Cell Biochemistry, Chinese Academy of Sciences). Luciferase activity was detected 48 hours later using the dual-luciferase reporter assay system (Promega, Madison, WI, USA) according to the manufacturer’s instructions. The experiment was repeated three times.
Isolation and Culture of Schwann Cells
The bilateral peripheral nerves (from the L5 spinal nerve to the division between the tibial nerve and common fibular nerve) were excised to 1 mm and stored in F12 medium at 4°C. After adding 5 mg/mL collagenase (Sigma-Aldrich, Merck KGaA, Darmstadt, Germany) and 1 mg/mL dispase II (Roche Diagnostics, Basel, Switzerland), nerves were incubated in phosphate-buffered saline (PBS) for 30 min. The cell suspension was gently dispersed with debris discarded through a cell strainer with a pore size of 60 μm (pluriSelect, GmbH, Leipzig, Germany). Next, cells were cultured in a 37°C incubator with 5% CO2 for 1 hour. Adherent cells were collected using 0.05% trypsin/ethylenediaminetetraacetic acid and then treated with Thy1.1 monoclonal antibody (Sigma-Aldrich) and rabbit complement (Sigma-Aldrich) to remove fibroblasts. Immunostaining against a specific marker, S100 (Sigma-Aldrich), was conducted to verify the Schwann cells.
Schwann Cell Grouping and Treatment
H19 lentivirus (LV)-based vector (LV-H19), siRNA-H19 and the corresponding negative controls were purchased from Gene Pharma (Shanghai, China). Then, 12 hours after LV-H19 or LV-NC infection, Schwann cells were treated with 30 μmol/L roscovitine (Selleck Chemicals, Houston, TX, USA) or treated with an equal volume of PBS for 15 hours. LV-H19 and LV-NC were transfected into Schwann cells according to the instructions for LipofectamineTM 2000 (Invitrogen, Carlsbad, CA, USA). Subsequent experiments were performed after 48 hours.
Enzyme-Linked Immunosorbent Assay (ELISA)
ELISA kits (Nanjing Jiancheng Bioengineering Institute, Nanjing, Jiangsu, China) were utilized to assess tumor necrosis factor (TNF-α), interleukin-1β (IL-1β) and IL-6 levels in homogenates of L4-L6 spinal dorsal horns.18
Reverse Transcription-Quantitative Polymerase Chain Reaction (RT-qPCR)
Total RNA from the L4-L5 spinal dorsal horn and cells was extracted by an RNA extraction kit (Takara, Dalian, China), and the RNA concentration was detected at a wavelength of 260 nm. Prime ScriptTM RT Master Mix (Takara) was used for reverse transcription, and SYBR Premix Ex Taq II (Takara) was used to perform qPCR. Primer sequences tested by Roche LightCycler 480 (Roche Diagnostics) are shown in Table 1. With the use of GAPDH expression for normalization, relative mRNA expression was calculated using the 2-ΔΔCt method.
 |
Table 1 Primers Sequence |
Western Blot Analysis
Total protein was extracted from the L4-L6 spinal dorsal horn and cells using a protein extraction kit (KeyGen Biotech., Ltd., Nanjing, Jiangsu, China), and the protein concentration was measured using the bicinchoninic acid method (Beyotime Biotechnology Co., Ltd, Nanjing, Jiangsu, China). The extracted proteins were separated by sodium dodecyl sulfate-polyacrylamide gel electrophoresis and transferred onto nitrocellulose filter membranes. The membranes were incubated with the following primary antibodies (all from Abcam Inc., Cambridge, MA, USA): rabbit anti-GABA B Receptor 2 (GABAB2) antibody (1:1000, ab52248), rabbit anti-glial fibrillary acidic protein (GFAP) antibody (1:2000, ab33922), rabbit anti-5HT2A receptor antibody (1: 300, ab216959), rabbit anti-CaMK II (phospho-T286) antibody (1:1000, ab32678), rabbit anti-CREB (phospho-S133) antibody (1:5000, ab32096), rabbit anti-CDK5 antibody (1:2000, ab40773), α-tubulin antibody (1:5000; ab7291), and β-actin antibody (1:5000; ab8226). The secondary antibody was horseradish peroxidase-labeled goat anti-rabbit immunoglobulin G (IgG) (1:5000, ab205718). Every protein band density was calculated with Image-Pro Plus 6.0 (Media Cybernetics, Silver Spring, USA).
Immunofluorescence Assay
The spinal dorsal horn tissues fixed with 4% paraformaldehyde were embedded in OCT compound after dehydration, frozen and sliced into sections of 15 μm. After permeabilization and fixation with precooled 100% formaldehyde, the sections were subjected to antigen retrieval and blocked with 5% serum for 30 min at room temperature. Then, the sections were incubated with anti-CDK5 (1:100, ab40773), anti-CREB (phospho-S133) (1:100, ab32096), anti-CaMKII (phospho-T286) (1:100, ab32678) or anti-NeuN (1:1000, ab104224) at 4°C overnight, followed by incubation with FITC-labeled goat anti-rabbit IgG (1:1000, ab6717) or Alexa Fluor® 647-labeled IgG (1:200, ab150115) away from light. The images were captured using a fluorescence microscope (ECLIPSE TE2000-S, Nikon Corporation Instruments Company, Japan).
Hematoxylin-Eosin (HE) Staining
Spinal dorsal horns were fixed with 4% paraformaldehyde, embedded in paraffin wax and sliced in sections of 6 mm. According to a standard protocol, the sections were stained by HE and observed under a light microscope.
Statistical Analysis
SPSS 21.0 (IBM Corp. Armonk, NY, USA) was used for data analysis. The Kolmogorov–Smirnov test indicated whether the data were normally distributed. The results are shown as the mean ± standard deviation. The t-test was used for analyzing comparisons between two groups, one-way analysis of variance (ANOVA) or two-way ANOVA was used for comparing multiple groups, and Tukey’s multiple comparisons test was used for pairwise comparisons after ANOVA. The p value was obtained using a two-tailed test, and p < 0.05 indicated a significant difference.
Results
Inflammatory Factors are Upregulated While 5-HT2A and GABAB2 are Decreased in NP Rats
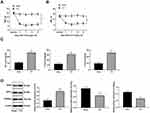
The NP rat model was successfully established with CCI surgery. The PWT and PWL of rats were both clearly decreased until 21 days after CCI surgery (Figure 1A and B) (both p < 0.01). In addition, inflammatory-related factors TNF-α, IL-1β and IL-6 in the rat spinal cord were upregulated after CCI surgery (Figure 1C) (all p < 0.01), and astrocyte cell-related protein GFAP levels were increased, while the suppressive neurotransmitter-related factors 5-HT2A and GABAB2 expression decreased (Figure 1D) (all p < 0.01).
NP Rats Have Increased CREB Phosphorylation via the CDK5/p35 Axis
In the NP rats established by CCI surgery, H19 expression in spinal dorsal horns was significantly promoted (Figure 2A) (p < 0.01). Accumulating evidence suggests that the activity of CDK5 is related to the expression of p35.21–23 Compared with those in the sham-operated rats, CDK5 and p35 protein levels in the NP rats were greatly enhanced, which was consistent with the results of the immunofluorescence assay (Figure 2B and C) (all p < 0.01). At the same time, CEEB and CaMK II phosphorylation levels were obviously increased (Figure 2D) (both p < 0.01). According to previous studies,24,25 p-CREB and p-CAMKII are mainly located in neurons. We obtained similar results using double immunofluorescence and found increased expression of p-CAMKII and p-CREB in neurons of NP rats.
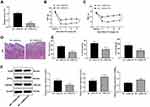
siRNA-H19 Alleviates NP in Rats
In the above study, we found that the expression of H19 in NP rats was significantly upregulated, so we interfered with the expression of H19 through siRNA and found that H19 expression in NP rats decreased after siRNA-H19 treatment (Figure 3A) (p < 0.01), while PWT and PWL strongly increased (Figure 3B and C) (both p < 0.01). HE staining showed that intrathecal injection of siRNA had little effect on spinal dorsal horn tissues since there was no obvious tissue deformation, necrosis or inflammatory cell infiltration (Figure 3D). Additionally, in NP rats, TNF-α, IL-1β, IL-6 and GFAP expression were downregulated, while 5HT2A and GABAB2 were upregulated, after siRNA-H19 treatment (Figure 3E and F) (all p < 0.01).
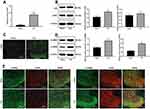
siRNA-H19 Downregulates CDK5/p35 to Reduce CREB Phosphorylation
In NP rats treated with siRNA-H19, the levels of CDK5 and p35 decreased, while by immunofluorescence, the expression of CDK5 decreased with decreasing H19 levels (Figure 4A and B, p < 0.05). With the decrease in H19 expression, the levels of p-CAMKII and p-CREB in the spinal cord of rats decreased significantly (Figure 4C, both p < 0.01).
siRNA-H19 and the CDK5 Inhibitor Roscovitine Decrease CDK5 and CREB Phosphorylation in vitro
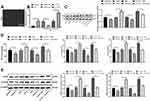
Schwann cells (Figure 5A) were isolated from rat spinal dorsal horns, and the results indicated that H19 expression in Schwann cells isolated from NP rats was much higher than that in cells from normal rats, and H19 was inhibited after transfection of siRNA-H19 and overexpressed after transfection with LV-H19 (Figure 5B) (both p < 0.01). Through detection assays, we found that siRNA-H19 reduced the levels of GFAP and inflammatory factors in Schwann cells isolated from NP rats, while cells overexpressing H19 had a reversed outcome. The CDK5 inhibitor roscovitine clearly reduced the levels of GFAP and inflammatory factors in Schwann cells. Additionally, the proinflammatory effect of H19 overexpression decreased when cells were treated with the CDK5 suppressor roscovitine (Figure 5C–E) (all p < 0.01).
More cells treated in an identical manner were collected to assess the changes in CDK5 and CREB phosphorylation. It was found that CDK5 and p-CREB expression decreased in cells transfected with siRNA-H19 but increased in cells overexpressing H19. However, roscovitine importantly reduced CDK5 and p-CREB expression in cells overexpressing H19 (Figure 5D) (all p < 0.01).
H19 Targets miR-196a-5p and Regulates CDK5 Expression
From the above experiments, we found that H19 affects the phosphorylation level of CREB by regulating CDK5. To further explore the specific mechanism by which H19 regulates CDK5, we took into consideration that lncRNA can regulate downstream mRNA expression through competitively binding to miR via the ceRNA mechanism.26 Therefore, through bioinformatics analysis, we found binding sites between H19 and miR-196a-5p and between miR-196a-5p and CDK5, which were further confirmed by dual-luciferase experiments (Figure 6A and B, p < 0.01). In addition, after knockdown of H19 expression in NP model rats and Schwann cells, the expression of miR-196a-5p was significantly upregulated, while the expression of CDK5 was downregulated. In addition, the expression of miR-196a-5p was significantly downregulated in Schwann cells overexpressing H19, while the expression of CDK5 was significantly upregulated (Figure 6C and D, p < 0.01).
Discussion
NP exerts lifelong unfavorable effects on patients, and since NP mechanisms are heterogeneous, and it is difficult to determine specific pain types, which may contribute to the poor treatment outcomes in this population.7 As an important kind of imprinted oncofetal lncRNA, H19 plays a major role in tumorigenesis and participates in almost every step in cancer cell progression by promoting cancer cell proliferation and cancer cell resistance to stress.27 In a previous study, it was discovered that in NP patients, H19 was consistently increased in Schwann cells from the peripheral axon in the primary sensory neurons, suggesting that H19 played a pivotal role in NP pathogenesis.10 In this study, we hypothesized that there is a mechanism of H19 in NP regulating CDK5 and CREB phosphorylation. Consequently, our data showed that silencing H19 mitigated NP by downregulating CDK5-mediated CREB phosphorylation.
NP rats presented decreased PWT and PWL. Both PWT and PWL are measures of thermal hyperalgesia and mechanical allodynia in the NP.28 Then, in NP rats, high H19 expression, CDK5 and CREB phosphorylation were observed. Interestingly, H19 activated inflammatory processes and was upregulated in Schwann cells, eventually leading to severe NP.10 CDK5 is closely linked with central nervous system and neuron activities because it controls memory improvement, synaptic functions and neuronal differentiation and migration.29 When mechanical allodynia and thermal hyperalgesia were induced by chronic dorsal root ganglia compression surgery, CREB, the protein related to pain, was greatly promoted.30
In addition, siRNA-H19 was found to have the ability to mitigate NP in model rats by downregulating TNF-α, IL-1β, IL-6 and GFAP levels while upregulating 5-HT2A and GABAB2. Growing evidence implies that inflammatory reactions induced by nerve injury in turn promote NP progression.31 As indicated in a prior study, H19 was positively correlated with TNF-α in ischemic stroke.32 Actively expressed H19 upregulated the proinflammatory factors IL-1β and IL-6 by promoting microglia and astrocyte activation.33 In NC model rats where substance P was injected, GFAP decreased as mechanical allodynia was mitigated; on the other hand, anti-inflammatory factors were actively expressed.34 Moreover, 5-HT2A has the ability to increase potassium chloride cotransporter type 2, thereby mediating motoneuronal inhibition and alleviating NP induced by spinal cord injury.35 GABAB2, which plays a role in maintaining the analgesic function of opioid oxycodone, was reduced with nerve injury.36
In our experiment, in vitro CDK5 and CREB phosphorylation was reduced and TNF-α, IL-1β, IL-6 and GFAP levels were inhibited, while 5-HT2A and GABAB2 were enhanced with the combination of siRNA-H19 and the CDK5 inhibitor roscovitine. Interestingly, Morales I and his colleagues indicated that microglial cells activated by endogenous injury released TNF-α, IL-1β and IL-6, inducing the signaling cascade and activating CDK5.37 In chronic ocular hypertension characterized by high GFAP expression, CREB levels were also enhanced.38 Furthermore, in rats with liver injury, 5-HT2A and GABAB2 directly repressed CREB expression, exerting protective effects on liver neurons.39 Increased CREB phosphorylation in the dorsal horn resulted in an increase in histone H4 acetylation in the CDK5 promoter and upregulated CDK5 transcription, so suppressed CREB expression reduced CDK5 growth and relieved rat mechanical allodynia.13 Importantly, it was previously indicated that dysregulated H19 can directly or indirectly modulate many different carcinogenic factors, including CDK.40 In pancreatic neuroendocrine neoplasms, H19, acting as an oncogene, upregulated CREB expression to induce cancer cell growth.41 Overall, our data showed that silencing H19 was helpful in blocking NP progression.
The ceRNA network of mRNA, lncRNA, and miR in diseases has been identified.42 Furthermore, we discovered that H19 competitively bound to miR-196a-5p to upregulate CDK5 expression. After knockdown of H19 expression in NP model rats and Schwann cells, miR-196a-5p expression was significantly upregulated, while CDK5 expression was downregulated. Early intervention of miR-196a delivered by an adeno-associated virus vector ameliorated the spinal and bulbar muscular atrophy phenotypes in a mouse model.14 Thus far, there is no study on the expression profiling and mechanism of miR-196a-5p in NP. Overall, silencing H19 inhibited NP by suppressing CDK5/p35 and p-CREB phosphorylation via the miR-196a-5p/CDK5 axis.
Conclusion
In summary, our study demonstrated that lncRNA H19 promoted NP by activating CDK5/p35-mediated CREB phosphorylation. These results identified a novel approach for NP treatment. In the future, we will further explore the underlying mechanism of other targets of lncRNA H19. More attention will be paid to seeking reliable therapeutic targets of NP. Nevertheless, this is solely preclinical research, and although our findings have therapeutic implications for NP treatment, the experimental results and effective application in clinical practice need further validation. There are many cell types involved in neuropathic pain in the spinal cord. Due to the limitations of experimental funding and experimental cycles, we mainly studied H19 in the spinal cord and Schwann cells. In the future, we will carry out a more in-depth investigation.
Abbreviations
LncRNA, long noncoding RNA; NP, neuropathic pain; CDK5, cyclin-dependent kinases 5; CREB, cAMP-response element binding protein; CCI, chronic constriction injury; PWT, paw withdrawal threshold; PWL, paw withdrawal latency; siRNA, small interfering RNA; LV, lentivirus; PBS, phosphate buffered saline; ELISA, enzyme-linked immunoSorbent assay; TNF-α, Tumor necrosis factor-alpha; IL-1β, concentrations of interleukin-1 beta; RT-qPCR, reverse transcription-quantitative polymerase chain reaction; IgG, immunoglobulin G; ANOVA, analysis of variance; GFAP, glial fibrillary acidic protein; NP, neuropathic pain; 5-HT2A, 5-hydroxytryptamine 2A; GABAB2, γ-Aminobutyric acid receptor 2; CaMK II, concentrations of interleukin-1 beta; IL-6, Interleukin- 6; H&E, hematoxylin and eosin.
Disclosure
The authors report no conflicts of interest for this work.
References
1. Finnerup NB, Haroutounian S, Kamerman P, et al. Neuropathic pain: an updated grading system for research and clinical practice. Pain. 2016;157(8):1599–1606. doi:10.1097/j.pain.0000000000000492
2. Gilron I, Baron R, Jensen T. Neuropathic pain: principles of diagnosis and treatment. Mayo Clin Proc. 2015;90(4):532–545. doi:10.1016/j.mayocp.2015.01.018
3. Watson JC, Sandroni P. Central neuropathic pain syndromes. Mayo Clin Proc. 2016;91(3):372–385. doi:10.1016/j.mayocp.2016.01.017
4. Cohen SP, Mao J. Neuropathic pain: mechanisms and their clinical implications. BMJ. 2014;348:f7656. doi:10.1136/bmj.f7656
5. Gierthmuhlen J, Baron R. Neuropathic pain. Semin Neurol. 2016;36(5):462–468. doi:10.1055/s-0036-1584950
6. Zilliox LA. Neuropathic pain. Continuum. 2017;23:512–532. doi:10.1212/CON.0000000000000462
7. Widerstrom-Noga E. Neuropathic pain and spinal cord injury: phenotypes and pharmacological management. Drugs. 2017;77(9):967–984. doi:10.1007/s40265-017-0747-8
8. Guo ZH, You ZH, Wang YB, Yi HC, Chen ZH. A learning-based method for LncRNA-disease association identification combing similarity information and rotation forest. iScience. 2019;19:786–795. doi:10.1016/j.isci.2019.08.030
9. Peng L, Yuan XQ, Liu ZY, et al. High lncRNA H19 expression as prognostic indicator: data mining in female cancers and polling analysis in non-female cancers. Oncotarget. 2017;8(1):1655–1667. doi:10.18632/oncotarget.13768
10. Iwasaki H, Sakai A, Maruyama M, Ito T, Sakamoto A, Suzuki H. Increased H19 long non-coding RNA expression in schwann cells in peripheral neuropathic pain. J Nippon Med Sch. 2019;86(4):215–221. doi:10.1272/jnms.JNMS.2018_86-402
11. Shupp A, Casimiro MC, Pestell RG. Biological functions of CDK5 and potential CDK5 targeted clinical treatments. Oncotarget. 2017;8(10):17373–17382. doi:10.18632/oncotarget.14538
12. Pozo K, Bibb JA. The emerging role of Cdk5 in cancer. Trends Cancer. 2016;2(10):606–618. doi:10.1016/j.trecan.2016.09.001
13. Li K, Zhao GQ, Li LY, Wu GZ, Cui SS. Epigenetic upregulation of Cdk5 in the dorsal horn contributes to neuropathic pain in rats. Neuroreport. 2014;25(14):1116–1121. doi:10.1097/WNR.0000000000000237
14. Miyazaki Y, Adachi H, Katsuno M, et al. Viral delivery of miR-196a ameliorates the SBMA phenotype via the silencing of CELF2. Nat Med. 2012;18(7):1136–1141. doi:10.1038/nm.2791
15. Wang GQ, Cen C, Li C, et al. Deactivation of excitatory neurons in the prelimbic cortex via Cdk5 promotes pain sensation and anxiety. Nat Commun. 2015;6:7660. doi:10.1038/ncomms8660
16. Barbosa S, Carreira S, Bailey D, Abaitua F, O’Hare P. Phosphorylation and SCF-mediated degradation regulate CREB-H transcription of metabolic targets. Mol Biol Cell. 2015;26(16):2939–2954. doi:10.1091/mbc.E15-04-0247
17. Wang L, Hu XH, Huang ZX, et al. Regulation of CREB functions by phosphorylation and sumoylation in nervous and visual systems. Curr Mol Med. 2017;16(10):885–892. doi:10.2174/1566524016666161223110106
18. Xie T, Zhang J, Kang Z, Liu F, Lin Z. miR-101 down-regulates mTOR expression and attenuates neuropathic pain in chronic constriction injury rat models. Neurosci Res. 2019. doi:10.1016/j.neures.2019.09.002
19. Betel D, Koppal A, Agius P, Sander C, Leslie C. Comprehensive modeling of microRNA targets predicts functional non-conserved and non-canonical sites. Genome Biol. 2010;11(8):R90. doi:10.1186/gb-2010-11-8-r90
20. Li JH, Liu S, Zhou H, Qu LH, Yang JH. StarBase v2.0: decoding miRNA-ceRNA, miRNA-ncRNA and protein-RNA interaction networks from large-scale CLIP-Seq data. Nucleic Acids Res. 2014;42:D92–97. doi:10.1093/nar/gkt1248
21. Amin ND, Zheng Y, Bk B, et al. The interaction of Munc 18 (p67) with the p10 domain of p35 protects in vivo Cdk5/p35 activity from inhibition by TFP5, a peptide derived from p35. Mol Biol Cell. 2016;27(21):3221–3232. doi:10.1091/mbc.E15-12-0857
22. Diaz A, Jeanneret V, Merino P, McCann P, Yepes M. Tissue-type plasminogen activator regulates p35-mediated Cdk5 activation in the postsynaptic terminal. J Cell Sci. 2019;132(5):jcs224196. doi:10.1242/jcs.224196
23. Kwon YT, Tsai LH, Crandall JE. Callosal axon guidance defects in p35(-/-) mice. J Comp Neurol. 1999;415(2):218–229. doi:10.1002/(sici)1096-9861(19991213)415:2<218::aid-cne6>3.0.co;2-f
24. Yao CY, Weng ZL, Zhang JC, Feng T, Lin Y, Yao S. Interleukin-17A acts to maintain neuropathic pain through activation of CaMKII/CREB signaling in spinal neurons. Mol Neurobiol. 2016;53(6):3914–3926. doi:10.1007/s12035-015-9322-z
25. Crown ED, Gwak YS, Ye Z, et al. Calcium/calmodulin dependent kinase II contributes to persistent central neuropathic pain following spinal cord injury. Pain. 2012;153(3):710–721. doi:10.1016/j.pain.2011.12.013
26. Zhong ME, Chen Y, Zhang G, Xu L, Ge W, Wu B. LncRNA H19 regulates PI3K-Akt signal pathway by functioning as a ceRNA and predicts poor prognosis in colorectal cancer: integrative analysis of dysregulated ncRNA-associated ceRNA network. Cancer Cell Int. 2019;19:148. doi:10.1186/s12935-019-0866-2
27. Raveh E, Matouk IJ, Gilon M, Hochberg A. The H19 Long non-coding RNA in cancer initiation, progression and metastasis - a proposed unifying theory. Mol Cancer. 2015;14:184. doi:10.1186/s12943-015-0458-2
28. Yu HM, Wang Q, Sun WB. Silencing of FKBP51 alleviates the mechanical pain threshold, inhibits DRG inflammatory factors and pain mediators through the NF-kappaB signaling pathway. Gene. 2017;627:169–175. doi:10.1016/j.gene.2017.06.029
29. Liu SL, Wang C, Jiang T, Tan L, Xing A, Yu JT. The role of Cdk5 in Alzheimer’s disease. Mol Neurobiol. 2016;53(7):4328–4342. doi:10.1007/s12035-015-9369-x
30. Xu F, Zhao X, Liu L, et al. Perturbing NR2B-PSD-95 interaction relieves neuropathic pain by inactivating CaMKII-CREB signaling. Neuroreport. 2017;28(13):856–863. doi:10.1097/WNR.0000000000000849
31. Liu S, Mi WL, Li Q, et al. Spinal IL-33/ST2 signaling contributes to neuropathic pain via neuronal CaMKII-CREB and astroglial JAK2-STAT3 cascades in mice. Anesthesiology. 2015;123(5):1154–1169. doi:10.1097/ALN.0000000000000850
32. Wang J, Zhao H, Fan Z, et al. Long noncoding RNA H19 promotes neuroinflammation in ischemic stroke by driving histone deacetylase 1-dependent M1 microglial polarization. Stroke. 2017;48(8):2211–2221. doi:10.1161/STROKEAHA.117.017387
33. Han CL, Ge M, Liu YP, et al. LncRNA H19 contributes to hippocampal glial cell activation via JAK/STAT signaling in a rat model of temporal lobe epilepsy. J Neuroinflammation. 2018;15(1):103. doi:10.1186/s12974-018-1139-z
34. Chung E, Yoon TG, Kim S, Kang M, Kim HJ, Son Y. Intravenous administration of substance P attenuates mechanical allodynia following nerve injury by regulating neuropathic pain-related factors. Biomol Ther. 2017;25(3):259–265. doi:10.4062/biomolther.2016.137
35. Sanchez-Brualla I, Boulenguez P, Brocard C, et al. Activation of 5-HT2A receptors restores KCC2 function and reduces neuropathic pain after spinal cord injury. Neuroscience. 2018;387:48–57. doi:10.1016/j.neuroscience.2017.08.033
36. Malcangio M. GABAB receptors and pain. Neuropharmacology. 2018;136:102–105. doi:10.1016/j.neuropharm.2017.05.012
37. Morales I, Farias G, Maccioni RB. Neuroimmunomodulation in the pathogenesis of Alzheimer’s disease. Neuroimmunomodulation. 2010;17(3):202–204. doi:10.1159/000258724
38. Gao F, Li F, Miao Y, et al. Involvement of the MEK-ERK/p38-CREB/c-fos signaling pathway in Kir channel inhibition-induced rat retinal Muller cell gliosis. Sci Rep. 2017;7(1):1480. doi:10.1038/s41598-017-01557-y
39. Shilpa J, Anju TR, Ajayan MS, Paulose CS. Increased cortical neuronal survival during liver injury: effect of gamma aminobutyric acid and 5-HT chitosan nanoparticles. J Biomed Nanotechnol. 2014;10(4):622–631. doi:10.1166/jbn.2014.1762
40. Wan X, Ding X, Chen S, et al. The functional sites of miRNAs and lncRNAs in gastric carcinogenesis. Tumour Biol. 2015;36(2):521–532. doi:10.1007/s13277-015-3136-5
41. Ji M, Yao Y, Liu A, et al. lncRNA H19 binds VGF and promotes pNEN progression via PI3K/AKT/CREB signaling. Endocr Relat Cancer. 2019;26(7):643–658. doi:10.1530/ERC-18-0552
42. Zhou H, Chen D, Xie G, Li J, Tang J, Tang L. LncRNA-mediated ceRNA network was identified as a crucial determinant of differential effects in periodontitis and periimplantitis by high-throughput sequencing. Clin Implant Dent Relat Res. 2020;22(3):424–450. doi:10.1111/cid.12911
 © 2020 The Author(s). This work is published and licensed by Dove Medical Press Limited. The full terms of this license are available at https://www.dovepress.com/terms.php and incorporate the Creative Commons Attribution - Non Commercial (unported, v3.0) License.
By accessing the work you hereby accept the Terms. Non-commercial uses of the work are permitted without any further permission from Dove Medical Press Limited, provided the work is properly attributed. For permission for commercial use of this work, please see paragraphs 4.2 and 5 of our Terms.
© 2020 The Author(s). This work is published and licensed by Dove Medical Press Limited. The full terms of this license are available at https://www.dovepress.com/terms.php and incorporate the Creative Commons Attribution - Non Commercial (unported, v3.0) License.
By accessing the work you hereby accept the Terms. Non-commercial uses of the work are permitted without any further permission from Dove Medical Press Limited, provided the work is properly attributed. For permission for commercial use of this work, please see paragraphs 4.2 and 5 of our Terms.