Back to Journals » Cancer Management and Research » Volume 12
Long-Noncoding RNA FGD5-AS1 Enhances the Viability, Migration, and Invasion of Glioblastoma Cells by Regulating the miR-103a-3p/TPD52 Axis
Authors Su D, Ji Z, Xue P, Guo S, Jia Q, Sun H
Received 11 March 2020
Accepted for publication 8 July 2020
Published 27 July 2020 Volume 2020:12 Pages 6317—6329
DOI https://doi.org/10.2147/CMAR.S253467
Checked for plagiarism Yes
Review by Single anonymous peer review
Peer reviewer comments 2
Editor who approved publication: Dr Eileen O'Reilly
Daoqing Su, Zhengang Ji, Pengfei Xue, Shengfu Guo, Qingbin Jia, Hanyu Sun
Department of Neurosurgery, Liaocheng People’s Hospital and Liaocheng Brain Hospital, Liaocheng Hospital Affiliated to Shandong First Medical University, Liaocheng City, Shandong Province, People’s Republic of China
Correspondence: Hanyu Sun Tel +86-18365751192
Email [email protected]
Purpose: This study was designed to explore the functional role of FYVE, RhoGEF, and PH domain containing 5 antisense RNA 1 (FGD5-AS1) and the underlying regulatory mechanism in the progression of glioblastoma (GBM).
Materials and Methods: FGD5-AS1 expression was analyzed in The Cancer Genome Atlas (TCGA), and then detected in GBM tissues and cells by quantitative reverse-transcription polymerase chain reaction. Viability, migration, and invasion of GBM cells were assessed using the MTT, wound healing, and transwell assays, respectively. StarBase/TargetScan analysis and dual-luciferase reporter gene (DLR) assay were performed to investigate the relationship between FGD5-AS1/tumor protein D52 (TPD52) and miR-103a-3p. A xenograft tumor model was established to evaluate the role of FGD5-AS1 in GBM tumorigenesis in vivo.
Results: FGD5-AS1 was overexpressed in GBM tissues and cells, and silencing of FGD5-AS1 expression resulted in the inhibition of the viability, migration, and invasion of GBM cells. miR-130-3p was a target of FGD5-AS1, and its expression was negatively regulated by FGD5-AS1. Silencing miR-103a-3p expression resulted in the abrogation of the inhibitory effects of si-FGD5-AS1 on the viability, migration, and invasion of GBM cells. TPD52 was a target of miR-103a-3p and suppressed the antitumor effects of FGD5-AS1 silencing on GBM cells. In addition, FGD5-AS1 silencing inhibited the growth of xenograft tumors in vivo by modulating the miR-103a-3p/TPD52 axis.
Conclusion: Silencing of FGD5-AS1 inhibited the viability, migration, and invasion of GBM cells by regulating the miR-103a-3p/TPD52 axis.
Keywords: glioblastoma, FGD5-AS1, viability, migration, invasion, miR-103a-3p, TPD52
Introduction
Glioblastoma (GBM) is a common and fatal malignant tumor of the central nervous system.1 The median survival of GBM patients using standard therapy is approximately 12–15 months, and the 5-year survival rate is only about 5% in the world.2 At present treatment for patients with GBM mainly includes surgical resection, radiotherapy, and chemotherapy.3–5 Evidence suggests that high cell proliferation, migration, and invasiveness are the major obstacles for the effective treatment of GBM.6 The invasive growth and dispersed nature of GBM together with high recurrence frequency contribute to poor disease prognosis.7 Therefore, it is imperative to explore the potential mechanism underlying GBM development and design novel therapeutic strategies for GBM treatment.
Long-noncoding RNAs (lncRNAs) are known to serve as pivotal regulators in the occurrence and development of malignant tumors,8,9 including GBM. For instance, the lncRNA HOTAIR functions as a regulator of cell cycle and proliferation in human GBM.10 In addition, the lncRNA MALAT1 is known to play the role of a tumor suppressor in the progression of GBM by downregulating the expression of matrix metalloproteinase 2 and inactivating extracellular signal-regulated kinase (ERK)/mitogen-activated protein kinase (MAPK) signaling.11 FYVE, RhoGEF, and PH domain containing 5 antisense RNA 1 (FGD5-AS1) has been found to be involved in several human malignancies. For instance, Ding et al12 speculated FGD5-AS1 as a plausible therapeutic target for colorectal cancer, and Liu et al13 revealed the ability of FGD5-AS1 to facilitate the progression of oral squamous cell carcinoma by targeting miR-520b. Moreover, FGD5-AS1 has been reported to accelerate the proliferation of non-small cell lung carcinoma (NSCLC) cells by regulating the hsa-miR-107/fibroblast growth factor receptor-like 1 (FGFRL1) axis.14 Nevertheless, the biological function and mechanism of FGD5-AS1 in GBM have been rarely studied.
Several lncRNAs have been shown to act as competitive endogenous RNAs (ceRNAs) and sponge certain microRNAs (miRNAs) in biological processes related to cancers such as GBM. For instance, SNHG1 contributes to the progression of glioma by sponging miR-194 to regulate the expression of pleckstrin homology like domain family A member 1 (PHLDA1).15 In addition, ZNFX1 antisense RNA 1 (ZFAS1) is known to promote the progression of GBM by modulating the miR-150-5p/proteolipid protein 2 (PLP2) axis,16 and miR-103a-3p is reported to contribute to tumor growth in many human cancers such as gastric cancer,17 oral squamous cell carcinoma,18 and colorectal cancer.19 In addition, a recent study showed that miR-103a-3p is a pivotal regulator in GBM progression.20 Yu et al21 discovered that linc00152 contributes to the progression of GBM stem cells via modulation of the miR-103a-3p/FEZ family zinc finger 1 (FEZF1)/cell division cycle 25A (CDC25A) axis. However, the relationship between FGD5-AS1 and miR-103a-3p in GBM is yet unclear.
Tumor protein D52 (TPD52) plays different roles in various types of malignancies22 and was recently found to be regulated by specific miRNAs implicated in cancer progression, including miR-224,23 miR-34a,24 and miR-218.25 Li et al24 found that miR-34a impedes cell migration and invasion in breast cancer by directly targeting TPD52. Further, Lu et al25 showed that highly upregulated in liver cancer (HULC) contributes to the progression of cervical cancer by modulating the miR-218/TPD52 axis. However, the relationship among FGD5-AS1, miR-103a-3p, and TPD52 is yet unknown.
In the present study, we first measured the expression level of FGD5-AS1 in GBM tissues and cells, and assessed the effect of FGD5-AS1 silencing on the viability, migration, and invasion of GBM cells. Next, the regulatory interactions among FGD5-AS1, miR-103a-3p, and TPD52 in GBM progression were investigated. Finally, an in vivo experiment was performed to validate the effect of FGD5-AS1 on GBM tumorigenesis. Our findings revealed a promising therapeutic target for GBM.
Materials and Methods
Tissue Samples
Tumor tissues were collected from 30 patients with GBM (16 male and 14 female; GBM group), and paired normal brain tissues were used as controls (Normal group). These patients were admitted to our hospital from January 2015 to December 2017, and did not receive radiotherapy or chemotherapy before surgery. Informed consent was obtained from all patients. This research was approved by the ethics committee of our hospital.
Cell Culture
Human GBM cell lines, A172, U87, U251, T98G, and LN229, and normal human astrocyte (NHA) cells were purchased from the American Type Culture Collection (ATCC, Manassas, VA, USA). All these cells have no pollution and mycoplasma contamination. All cell lines were cultured in Dulbecco’s modified Eagle’s medium (DMEM; GIBCO, Carlsbad, CA, USA) supplemented with 10% (v/v) fetal bovine serum (FBS) (GIBCO) in a humidified incubator at 37°C.
Cell Transfection
U251 and U87 cells were seeded in 24-well plates at a density of 1 × 105 cells/well and maintained for 24 h at 37°C. Small-interfering RNAs (siRNAs) targeting FGD5-AS1 (si-FGD5-AS1-1 and si-FGD5-AS1-2) and the corresponding negative control (si-NC) were obtained from GenePharma Co., Ltd (Shanghai, China). An overexpression plasmid of miR-103a-3p (miR-103a-3p) and negative control (miR-NC) as well as an miR-103a-3p inhibitor (miR inhibitor) and a negative control (inhibitor-NC) were obtained from GenePharma. U251 and U87 cells were infected with the oligonucleotides stated above using Lipofectamine 2000 (Invitrogen, Carlsbad, CA, USA). Cells without transfection were regarded as the blank group. To investigate the interaction between FGD5-AS1-1 and miR-103a-3p, U251 and U87 cells were co-transfected with si-FGD5-AS1 or si-NC and miR inhibitor or inhibitor-NC using Lipofectamine 2000.
pcDNA-TPD52 (TPD52) and negative control (pcDNA-NC) were purchased from GenePharma. To explore whether TPD52 was involved in the role of FGD5-AS1 in GBM, U251 cells were co-transfected with si-FGD5-AS1 or si-NC and pcDNA-NC or pcDNA-TPD52 using Lipofectamine 2000. Cells were analyzed after 48 h of transfection.
Quantitative Real-Time Polymerase Chain Reaction (qRT-PCR)
Total RNA was extracted from GBM cells and tissues using an miRNeasy Mini Kit (Qiagen, Duesseldorf, Germany), and 5 μL RNA was reverse transcribed into complementary DNA (cDNA) using Takara PrimeScript RT reagent kit with gDNA Eraser (Takara, Otsu, Japan). qRT-PCR was performed on an ABI 7500HT Fast Real-Time PCR System (Applied Biosystems, Waltham, MA, USA) under the following reaction conditions: an initial step at 95°C for 10 min, followed by 40 cycles of 95°C for 10 s, 60°C for 20 s, and 72°C for 34 s. The 2−ΔΔCT method was used to analyze data. GAPDH and U6 were used for normalization. The primer sequences used were as follows: lncRNA FGD5-AS1, 5′-CGTGGAGAAGAATTGGGC-3′ (forward) and 5′-CGTGGAGAAGAATTGGGC-3′ (reverse); miR-103a-3p, 5′-AGCAGCATTGTACAGGGCTATG-3′ (forward) and 5′-CTCTACAGCTATATTGCCAGCCAC-3′ (reverse); TPD52, 5′-GAGGAAGGAGAAGATGTTGC-3′ (forward) and 5′-GCCGAATTCAAGACTTCTCC-3′ (reverse); GAPDH, 5′-CCAGGTGGTCTCCTCTGACTT-3′ (forward) and 5′-GTTGCTGTAGCCAAATTCGTTGT-3′ (reverse); U6, 5′-CTCGCTTCGGCAGCACA-3′ (forward) and 5′-AACGCTTCACGAATTTGCGT-3′ (reverse).
Dual-Luciferase Reporter Gene (DLR) Assay
The binding site between miR-103a-3p and FGD5-AS1 was predicted by StarBase, while that between miR-103a-3p and TPD52 was predicted by TargetScan. The sequences of FGD5-AS1 WT and TPD52 WT were synthetized in accordance with the binding site and the corresponding sequences of FGD5-AS1 Mut and TPD52 Mut were synthetized on the basis of FGD5-AS1 WT and TPD52 WT. FGD5-AS1 WT/Mut and TPD52 WT/Mut were inserted into a pmirGLO dual-luciferase vector (Promega, Madison, WI, USA) to generate pmirGLO-FGD5-AS1 WT/Mut and pmirGLO-TPD52 WT/Mut, respectively. U251 and U87 cells were co-transfected with pmirGLO-FGD5-AS1 WT/Mut and miR-103a-3p mimics/mimics NC, or co-transfected with pmirGLO-TPD52 WT/Mut and miR-103a-3p mimics/mimics NC/miR-103a-3p + FGD5-AS1 using Lipofectamine 2000. After 48 h, the DLR assay system (Promega) was used to detect luciferase activity.
3-(4,5-Dimethyl-2-Thiazolyl)-2,5-Diphenyl-2-H-Tetrazolium Bromide (MTT) Assay
Cells were cultured in 96-well plates at a density of 6 × 103 cells/well for 0, 24, 48, and 72 h. Following incubation, 20 μL MTT reagent (5 mg/mL, Sigma-Aldrich, St. Louis, MO, USA) was added into each well and the plates were incubated for 4 h. Next, 150 μL dimethyl sulfoxide (DMSO) was added to terminate the reaction. Absorbance at 450 nm (A450) was measured using a microplate reader (Applied Biosystems).
Wound Healing Assay
Cells were seeded into six-well plates (5 × 105 cells/well) and an artificial scratch was created using a 10 μL pipette tip once they reached 90% confluency. Cells were cultured for 24 h and then observed under an inverted microscope (Olympus). Wound healing rate was calculated from the fraction of cell coverage across the line.
Transwell Invasion Analysis
Cell invasion was examined using transwell membranes (BD, Franklin Lakes, NJ, USA) coated with Matrigel and a serum-free medium. First, 1 × 105 cells were cultured in the upper chamber in 100 μL medium, while 500 μL medium containing 10% FBS was added as a chemoattractant to the lower chamber. After incubation for 48 h, invasive cells were stained with crystal violet for 10 min. Positively stained cells at five random fields were counted under a microscope (Olympus).
Xenograft Tumor Model
All experimental procedures were conducted according to the Chinese legislation regarding experimental animals. Male nude mice (BALB/c), 4-weeks-old, were obtained from HFK Bioscience Co., Ltd (Beijing, China). Mice were randomly divided into three groups as follows: Blank, si-NC, and si-FGD5-AS1 (N = 5 mice each group). U251 cells transfected with si-FGD5-AS1/si-NC (1 × 106 cells/nude mice) were subcutaneously injected into the left axilla of mice. The longest (L) and shortest (W) diameters of xenografted tumors were measured using a Vernier caliper every 7 days after injection. Tumor volume was calculated using the following formula: V = L × W2/2. After the last measurement on week 4 post-injection, mice were anesthetized by an intraperitoneal injection of sodium pentobarbital (80 mg/kg) and sacrificed by cervical dislocation. Xenografted tumors were completely excised and weighed. The expression of FGD5-AS1, miR-103a-3p, and TPD52 in xenografted tumors was detected by qRT-PCR as mentioned above.
Statistical Analysis
All assays were conducted at least three times and all data were analyzed using SPSS 22.0 statistical software and GraphPad Prism 7.0. Data are presented as means ± standard deviation (SD). Student’s t-test was performed to compare significant differences between two groups, while a one-way analysis of variance (ANOVA) followed by Tukey’s post hoc test was used to compare more than two groups. Differences were regarded statistically significant at P < 0.05.
Results
The lncRNA FGD5-AS1 Was Overexpressed in GBM
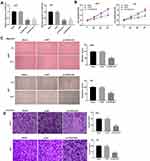
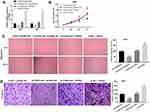
To study the functional role of FGD5-AS1 in GBM, we first analyzed FGD5-AS1 expression in The Cancer Genome Atlas (TCGA) and found it to be upregulated in GBM tissues (n = 163) as compared to that in normal tissues (n = 207) (P < 0.05, Figure 1A). To validate the data obtained from TCGA, the expression of FGD5-AS1 was analyzed in 30 GBM tissues and paired normal tissues by qRT-PCR. FGD5-AS1 expression was markedly elevated in GBM tissues than in normal tissues (P < 0.001, Figure 1B). We investigated the relationship between the expression of FGD5-AS1 and clinicopathological features of patients with GBM. As shown in Table 1, no association was observed between FGD5-AS1 expression and age, gender, tumor diameter, and resection degree (Table 1). Further, the expression of FGD5-AS1 was found to be upregulated in GBM cell lines (U251, L229, U87, T98G, and A172) as compared with that in NHAs (P < 0.01, Figure 1C). U251 and U87 cells with FGD5-AS1 expression relatively higher than other cell lines were used for further experiments.
 |
Table 1 The Relationship Between FGD5-AS1 Expression and Clinicopathological Features of Patients with Glioblastoma (GBM) |
 |
Figure 1 The expression of FGD5-AS1 in glioblastoma (GBM) tissues and cells. (A) Relative expression of FGD5-AS1 in GBM tissues and normal tissues based on The Cancer Genome Atlas (TCGA) (http://gepia.cancer-pku.cn/index.html). (B) Quantitative real-time polymerase chain reaction (qRT-PCR) was employed to assess the expression of FGD5-AS1 in 30 paired GBM tissues and normal tissues. (C) qRT-PCR was employed to assess the expression of FGD5-AS1 in GBM cell lines and normal human astrocyte (NHA) cells. *P < 0.05 vs Normal (A); **P < 0.01 vs NHA (C). |
Silencing of FGD5-AS1 Expression Impeded the Proliferation, Migration, and Invasion of GBM Cells
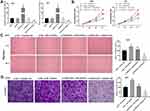
To evaluate the effect of FGD5-AS1 silencing on the progression of GBM, the expression of FGD5-AS1 was silenced by transfection of si-FGD5-AS1 into U251 and U87 cells. RT-qPCR revealed the remarkable decrease in FGD5-AS1 expression level in si-FGD5-AS1-1/2 groups as compared with that in the blank group (P < 0.01, Figure 2A). Transfection of si-NC had no effect on FGD5-AS1 expression. Considering the better inhibitory effects observed with si-FGD5-AS1-1, we used this siRNA for subsequent experiments. As shown in Figure 2B–D, the viability, migration, and invasion of cells remarkably declined following si-FGD5-AS1 treatment as compared with the effects observed in the blank group (P < 0.01). No remarkable difference was noticed between si-NC and blank groups. Taken together, silencing of FGD5-AS1 expression resulted in a decrease in the viability, migration, and invasion of GBM cells.
FGD5-AS1 Acted as a Sponge for miR-130-3p in GBM Cells
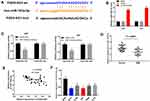
We used the StarBase online database and found miR-103a-3p as a potential target gene for FGD5-AS1. The binding site was found to be in the 3′-untranslated region (UTR) of FGD5-AS1 (Figure 3A). The expression of miR-103a-3p was found to be upregulated in si-FGD5-AS1–transfected U251 and U87 cells as compared with that in the blank group (P < 0.01, Figure 3B). To validate the target relationship between FGD5-AS1 and miR-103a-3p, a DLR assay was performed using U87 and U251 cells. We found that miR-103a-3p mimics markedly declined the luciferase activity of FGD5-AS1 WT but had no influence on the luciferase activity of FGD5-AS1 Mut (P < 0.01, Figure 3C). We also found that miR-103a-3p expression was downregulated in GBM tissues (P < 0.001, Figure 3D). Spearman correlation analysis revealed an inverse association between the relative expression of FGD5-AS1 and miR-103a-3p (r = −0.4224, P < 0.01, Figure 3E). Further, miR-103a-3p expression was also downregulated in GBM cell lines (P < 0.01, Figure 3F). Taken together, FGD5-AS1 acted as a sponge and reversely modulated the expression of miR-130-3p in GBM cells.
Inhibition of miR-103a-3p Expression Abrogated the Effect of FGD5-AS1 Silencing on the Viability, Migration, and Invasion of GBM Cells
To evaluate the biological role of miR-103a-3p in GBM, we overexpressed miR-103a-3p by transfection with miR-103a-3p or downregulated its expression using an miR-103a-3p inhibitor. qRT-PCR revealed a marked elevation in miR-103a-3p expression in miR-103a-3p treatment group and a consistent decrease in miR-103a-3p inhibitor group as compared with that in the blank group (P < 0.01, Figure 4A). To study the mechanism underlying the effects of FGD5-AS1 and miR-103a-3p in GBM cells, si-NC or si-FGD5-AS1 was co-transfected with miR-103a-3p inhibitor or inhibitor NC into U251 and U87 cells. The results of MTT analysis showed that cell viability markedly increased in si-NC + miR inhibitor group but significantly declined in si-FGD5-AS1 + inhibitor NC group as compared with that in si-NC + inhibitor NC group (P < 0.01, Figure 4B). In comparison with si-NC + inhibitor NC group, si-NC + miR inhibitor group showed a marked increase in the relative migration and invasion of cells, which remarkably decreased in si-FGD5-AS1 + inhibitor NC group (P < 0.01; Figure 4C and D). Treatment with miR-103a-3p inhibitor resulted in the abrogation of the inhibitory effect of si-FGD5-AS1 on the viability, migration, and invasion of GBM cells.
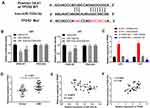
miR-103a-3p Directly Targeted TPD52
To reveal the mechanism underlying the function of miR-103a-3p in GBM, we performed TargetScan analysis to explore the target genes of miR-103a-3p. A binding site was predicted between miR-103a-3p and TPD52 by TargetScan (Figure 5A). DLR assay showed that miR-103a-3p expression remarkably decreased the luciferase activity of TPD52 WT, and this effect was partially reversed by FGD5-AS1 (P < 0.05, Figure 5B). To study the relationship between FGD5-AS1, miR-103a-3p, and TPD52, we co-transfected U251 and U87 cells with si-NC/si-FGD5-AS1 and inhibitor NC/miR-103a-3p inhibitor (miR inhibitor). The expression of TPD52 was markedly upregulated in si-NC + miR inhibitor group but significantly decreased in si-FGD5-AS1 + inhibitor NC group as compared with that in si-NC + inhibitor NC group. si-FGD5-AS1 reversed the effect of miR inhibitor on TPD52 expression (P < 0.01, Figure 5C). qRT-PCR indicated the overexpression of TPD52 in GBM tissues as compared with that in normal tissues (P < 0.001, Figure 5D). As shown in Figure 5E and F, TPD52 expression was negatively associated with miR-103a-3p expression (r = −0.3777, P < 0.01) and FGD5-AS1 and TPD52 expression showed a positive association (r = 0.3069, P < 0.05). Further, miR-103a-3p could directly target TPD52 and the effect of miR-103a-3p expression inhibition on the level of TPD52 was partially counteracted by FGD5-AS1 silencing.
Overexpression of TPD52 Partly Eliminated the Action of FGD5-AS1 Silencing on the Viability, Migration, and Invasion of GBM Cells
Considering that TPD52 is a target gene of miR-103a-3p, the effect of miR-103a-3p inhibition on TPD52 expression was partially counteracted by FGD5-AS1 silencing. To study whether TPD52 participates in the effect mediated by FGD5-AS1 in GBM, U251 and U87 cells were co-transfected with si-FGD5-AS1 or si-NC and pcDNA-TPD52 (TPD52) or pcDNA-NC. As shown in Figure 6A, TPD52 expression was downregulated in si-FGD5-AS1 + pcDNA-NC group but markedly increased in si-NC + TPD52 group as compared with that in si-NC + pcDNA-NC group (P < 0.01). The inhibitory effect of si-FGD5-AS1 on the expression of TPD52 was partly reversed by pcDNA-TPD52. As shown in Figure 6B–D, cell viability, migration, and invasion rate markedly decreased in si-FGD5-AS1 + pcDNA-NC group but significantly increased in si-NC + TPD52 group (P < 0.01). Further, pcDNA-TPD52 partly eliminated the inhibitory effect of si-FGD5-AS1 on the viability, migration, and invasion of U251 cells. Thus, silencing of FGD5-AS1 expression may result in the inhibition of the viability, migration, and invasion of U251 cells through the downregulation of TPD52 expression.
Silencing of FGD5-AS1 Expression Suppressed GBM Tumor Growth in vivo
To explore the effects of FGD5-AS1 expression on GBM tumorigenesis in vivo, U251 cells transfected with si-FGD5-AS1 were subcutaneously injected into mice. As presented in Figure 7A and B, tumor volumes and weights remarkably decreased in si-FGD5-AS1 group as compared with those in the blank group (P < 0.05). Further, a significant decrease in the expression of both FGD5-AS1 and TPD52 was observed, consistent with a remarkable increase in the expression of miR-103a-3p in si-FGD5-AS1 group as compared with that in the blank group (P < 0.01, Figure 7C). Transfection of si-NC had no effect on tumor volume, tumor weight, and FGD5-AS1, miR-103a-3p, and TPD52 expression as compared with the blank group. Taken together, silencing of FGD5-AS1 expression resulted in the suppression of GBM tumor growth in vivo through the modulation of the miR-103a-3p/TPD52 axis.
Discussion
Recent evidence suggests the critical role of lncRNAs in the progression and development of human malignancies, including GBM. Here, we studied the biological function of FGD5-AS1 and its mechanism of action in GBM. FGD5-AS1 is associated with cancer progression; it is overexpressed in colorectal cancer tissues and cells, wherein knockdown of FGD5-AS1 expression resulted in the repression of cell proliferation, migration, and invasion and induction of apoptosis.12 Fan et al14 revealed the overexpression of FGD5-AS1 in NSCLC, where it promotes cell proliferation. In the present study, the effect of FGD5-AS1 on the progression of GBM was in line with that reported in the abovementioned cancers. We found that FGD5-AS1 expression was upregulated in GBM tissues and cells and that FGD5-AS1 silencing could suppress the viability, migration, and invasion of GBM cells. Our findings reveal the potential oncogenic role of FGD5-AS1 in GBM.
Studies have shown that lncRNAs can act as ceRNAs and modulate the expression of their target genes by sponging miRNAs.26–28 Here, we demonstrate that miR-103a-3p, found to be dysregulated in various human tumors such as osteosarcoma tissues (upregulation)29 and glioma (downregulation),21 is a target gene of FGD5-AS1. Our data show that miR-103a-3p expression was downregulated in GBM tissues, as reported in previous studies. miR-103a-3p is thought to exert antitumor effects in different tumors such as bladder cancer30 and glioma.31 Notably, miR-103a-3p may act as a target gene for many lncRNAs in human tumors. Tang et al32 found that AFAP1-AS1 functions as a ceRNA for miR-103a-3p and promotes the proliferation of pituitary adenoma cells. Yu et al21 also showed the linc00152-mediated malignant progression of glioma stem cells via sponging of miR-103a-3p. However, the relationship between FGD5-AS1 and miR-130-3p in GBM is yet unclear. Here, we found that miR-130-3p is a target gene of FGD5-AS1 and that silencing miR-103a-3p expression resulted in the abrogation of the inhibitory effects of si-FGD5-AS1 on the viability, migration, and invasion of GBM cells. Our data reveal that the silencing of FGD5-AS1 expression may result in the inhibition of the progression of GBM through the sponging of miR-103a-3p.
TPD52 is a coiled-coil motif-bearing protein that plays a regulatory role in tumor progression. Previous studies have shown the upregulated expression of TPD52 and its function as a tumor promoter in prostate cancer,33 colorectal cancer,34 lung squamous cell carcinoma,35 and breast cancer.24 Dasari et al33 showed that TPD52 overexpression promotes the migration and proliferation of prostate cancer cells, while Kumamoto et al35 revealed the suppressive effect of TPD52 knockdown on the migration and invasion of lung squamous cell carcinoma cells. On the contrary, TPD52 is also identified as a tumor suppressor in renal cell carcinoma and hepatocellular carcinoma, as downregulation of TPD52 expression was associated with the poor prognosis of hepatocellular carcinoma.36 Zhao et al37 demonstrated the inhibitory effect of TPD52 overexpression on the proliferation, migration, and invasion of renal cell carcinoma cells. However, the specific regulatory role of TPD52 in GBM is yet questionable. We, here, show that TPD52 expression was upregulated in GBM tissues, consistent with its expression in prostate cancer, colorectal cancer, lung squamous cell carcinoma, and breast cancer. Our findings indicate that TPD52 may function as an oncogene in GBM. As the function of TPD52 is different in different cancers, further research is warranted on the regulatory effect and mechanism of action of TPD52 in different cancers.
TPD52 acts as a target of several miRNAs in different tumors. For instance, Han et al38 showed that miR-218 suppresses cell growth and facilitates cell apoptosis in prostate cancer by inhibiting TPD52 expression. Zhang et al39 revealed the anti-proliferative and apoptotic effects of miR-224-5p on GBM cells mediated via the downregulation of TPD52 expression. In line with these studies, we show that TPD52 is a target of miR-103a-3p. TPD52 expression negatively correlated with FGD5-AS1 level and TPD52 prevented the antitumor effects of FGD5-AS1 silencing on U251 cells. Thus, the silencing of FGD5-AS1 expression may result in the inhibition of the progression of GBM through the regulation of the miR-103a-3p/TPD52 axis. The regulatory mechanism associated with the FGD5-AS1/miR-103a-3p/TPD52 axis was further validated in our in vivo experiments.
This study has a few limitations. First, the regulatory effects of the FGD5-AS1/miR-103a-3p/TPD52 axis on GBM stem cells were not evaluated. GBM stem cell may be a better representative of human GBM, and the detection of protein expression of TPD52 in GBM stem cell lines may be reflective of the regulatory role of this protein in GBM. Second, the downstream signaling pathways associated with the FGD5-AS1/miR-103a-3p/TPD52 axis in GMB are still unclear. Third, the small clinical samples lead to a possible statistical error, and further studies with larger number of samples are warranted to confirm these results.
Conclusions
FGD5-AS1 expression was upregulated in GBM tissues and cells, and silencing of FGD5-AS1 expression resulted in the suppression of the viability, migration, and invasion of GBM cells through the regulation of the miR-130-3p/TPD52 axis. Further, FGD5-AS1 silencing also inhibited the growth of GBM tumor in vivo. Our findings highlight the FGD5-AS1/miR-103a-3p/TPD52 axis as a potential therapeutic target for GBM.
Ethics Approval and Consent to Participate
This study was approved by the ethics committee of Liaocheng People’s Hospital and Liaocheng Brain Hospital, Liaocheng Hospital Affiliated To Shandong First Medical University. Written informed consent was obtained from all subjects.
Animal experiments were performed in accordance with the “Guidelines for the Care and Use of Laboratory Animals” published by the National Institutes of Health and were approved by Liaocheng People’s Hospital and Liaocheng Brain Hospital, Liaocheng Hospital Affiliated To Shandong First Medical University’s animal care and use committee.
Author Contributions
All authors made substantial contributions to conception and design, acquisition of data, or analysis and interpretation of data; took part in drafting the article or revising it critically for important intellectual content; gave final approval of the version to be published; and agree to be accountable for all aspects of the work.
Disclosure
The authors report no funding and no conflicts of interest for this work.
References
1. Delgado-López PD, Corrales-García EM. Survival in glioblastoma: a review on the impact of treatment modalities. Clin Transl Oncol. 2016;18(11):1062–1071. doi:10.1007/s12094-016-1497-x
2. Linder B, Weirauch U, Ewe A, et al. Therapeutic targeting of stat3 using lipopolyplex nanoparticle-formulated siRNA in a syngeneic orthotopic mouse glioma model. Cancers. 2019;11(3):333. doi:10.3390/cancers11030333
3. Kuhnt D, Becker A, Ganslandt O, Bauer M, Buchfelder M, Nimsky C. Correlation of the extent of tumor volume resection and patient survival in surgery of glioblastoma multiforme with high-field intraoperative MRI guidance. Neuro-Oncology. 2011;13(12):1339–1348. doi:10.1093/neuonc/nor133
4. Oppenlander ME, Wolf AB, Snyder LA, et al. An extent of resection threshold for recurrent glioblastoma and its risk for neurological morbidity. J Neurosurg. 2014;120(4):846–853. doi:10.3171/2013.12.JNS13184
5. Wu F, Zhang C, Cai J, et al. Upregulation of long noncoding RNA HOXA-AS3 promotes tumor progression and predicts poor prognosis in glioma. Oncotarget. 2017;8(32):53110–53123. doi:10.18632/oncotarget.18162
6. Ghosh D, Nandi S, Bhattacharjee S. Combination therapy to checkmate glioblastoma: clinical challenges and advances. Clin Transl Med. 2018;7(1):33. doi:10.1186/s40169-018-0211-8
7. Wen PY, Reardon DA. Neuro-oncology in 2015: progress in glioma diagnosis, classification and treatment. Nat Rev Neurol. 2016;12(2):69–70. doi:10.1038/nrneurol.2015.242
8. Li Y, Wang Z, Wang Y, et al. Identification and characterization of lncRNA mediated transcriptional dysregulation dictates lncRNA roles in glioblastoma. Oncotarget. 2016;7(29):45027–45041. doi:10.18632/oncotarget.7801
9. Liu S, Mitra R, Zhao MM, et al. The potential roles of long non-coding RNAs (lncRNAs) in glioblastoma development. Mol Cancer Ther. 2016;15(12):2977–2986. doi:10.1158/1535-7163.MCT-16-0320
10. Kailiang Z, Xiaotian S, Xuan Z, et al. Long non-coding RNA HOTAIR promotes glioblastoma cell cycle progression in an EZH2 dependent manner. Oncotarget. 2015;6(1):537–546. doi:10.18632/oncotarget.2681
11. Han Y, Wu Z, Wu T, et al. Tumor-suppressive function of long noncoding RNA MALAT1 in glioma cells by downregulation of MMP2 and inactivation of ERK/MAPK signaling. Cell Death Dis. 2016;7(3):e2123. doi:10.1038/cddis.2015.407
12. Li D, Jiang X, Zhang X, Cao G, Wang D, Chen Z. Long noncoding RNA FGD5-AS1 promotes colorectal cancer cell proliferation, migration, and invasion through upregulating CDCA7 via sponging miR-302e. In Vitro Cell Dev Biol Anim. 2019;55(8):577–585. doi:10.1007/s11626-019-00376-x
13. Liu L, Zhan Y, Huang Y, Huang L. LncRNA FGD5-AS1 can be predicted as therapeutic target in oral cancer. J Oral Pathol Med. 2020;3(10):12989.
14. Fan Y, Li H, Yu Z, et al. Long non-coding RNA FGD5-AS1 promotes non-small cell lung cancer cell proliferation through sponging hsa-miR-107 to up-regulate FGFRL1. Biosci Rep. 2020;40(1):854–859. doi:10.1042/BSR20193309
15. Liu L, Shi Y, Shi J, et al. The long non-coding RNA SNHG1 promotes glioma progression by competitively binding to miR-194 to regulate PHLDA1 expression. Cell Death Dis. 2019;10(6):463. doi:10.1038/s41419-019-1698-7
16. Li X, Luo Y, Liu L, Cui S, Luo L. The long noncoding RNA ZFAS1 promotes the progression of glioma by regulating the miR150-5p/PLP2 axis. J Cell Physiol. 2019;235(2):2937–2946. doi:10.1002/jcp.29199
17. Hu X, Miao J, Zhang M, et al. miRNA-103a-3p promotes human gastric cancer cell proliferation by targeting and suppressing ATF7 in vitro. Mol Cells. 2018;41(5):390–400. doi:10.14348/molcells.2018.2078
18. Zhang G, Chen Z, Zhang Y, Li T, Bao Y, Zhang S. Inhibition of miR-103a-3p suppresses the proliferation in oral squamous cell carcinoma cells via targeting RCAN1. Neoplasma. 2020;190430N190382.
19. Zheng Y-B, Xiao K, Xiao G-C, et al. MicroRNA-103 promotes tumor growth and metastasis in colorectal cancer by directly targeting LATS2. Oncol Lett. 2016;12(3):2194–2200. doi:10.3892/ol.2016.4814
20. Vastrad B, Vastrad C, Godavarthi A, Chandrashekar R. Molecular mechanisms underlying gliomas and glioblastoma pathogenesis revealed by bioinformatics analysis of microarray data. Med Oncol. 2017;34(11):182. doi:10.1007/s12032-017-1043-x
21. Yu M, Xue Y, Zheng J, et al. Linc00152 promotes malignant progression of glioma stem cells by regulating miR-103a-3p/FEZF1/CDC25A pathway. Mol Cancer. 2017;16(1):110. doi:10.1186/s12943-017-0677-9
22. Byrne JA, Frost S, Chen Y, Bright RK. Tumor protein D52 (TPD52) and cancer—oncogene understudy or understudied oncogene? Tumor Biol. 2014;35(8):7369–7382. doi:10.1007/s13277-014-2006-x
23. Goto Y, Nishikawa R, Kojima S, et al. Tumour-suppressive microRNA-224 inhibits cancer cell migration and invasion via targeting oncogenic TPD52 in prostate cancer. FEBS Lett. 2014;588(10):1973–1982. doi:10.1016/j.febslet.2014.04.020
24. Li G, Yao L, Zhang J, et al. Tumor-suppressive microRNA-34a inhibits breast cancer cell migration and invasion via targeting oncogenic TPD52. Tumor Biol. 2016;37(6):7481–7491. doi:10.1007/s13277-015-4623-4
25. Lu W, Wan X, Tao L, Wan J. Long non-coding RNA HULC promotes cervical cancer cell proliferation, migration and invasion via miR-218/TPD52 axis. Onco Targets Ther. 2020;13:1109–1118. doi:10.2147/OTT.S232914
26. Huang P, Huang FZ, Liu HZ, Zhang TY, Yang MS, Sun CZ. LncRNA MEG3 functions as a ceRNA in regulating hepatic lipogenesis by competitively binding to miR-21 with LRP6. Metabolism. 2019;94:1–8. doi:10.1016/j.metabol.2019.01.018
27. Liu L, Cui S, Wan T, Li X, Shi Y. Long non-coding RNA HOTAIR acts as a competing endogenous RNA to promote glioma progression by sponging miR-126-5p. J Cell Physiol. 2018;233(5):6822–6831. doi:10.1002/jcp.26432
28. Xia L, Nie D, Wang G, Sun C, Chen G. FER1L4/miR-372/E2F1 works as a ceRNA system to regulate the proliferation and cell cycle of glioma cells. J Cell Mol Med. 2019;23(5):3224–3233. doi:10.1111/jcmm.14198
29. Wang X, Lin Y, Peng L, et al. MicroRNA-103 promotes proliferation and inhibits apoptosis in spinal osteosarcoma cells by targeting p57. Oncol Res. 2018;26(6):933–940. doi:10.3727/096504017X15144741233346
30. Zhong Z, Lv M, Chen J. Screening differential circular RNA expression profiles reveals the regulatory role of circTCF25-miR-103a-3p/miR-107-CDK6 pathway in bladder carcinoma. Sci Rep. 2016;6(1):30919. doi:10.1038/srep30919
31. Kanayama S, Kaito T, Kitaguchi K, Ishiguro H, Yoshikawa H. ONO-1301 enhances in vitro osteoblast differentiation and in vivo bone formation induced by bone morphogenetic protein. Spine. 2017;43(11):1.
32. Tang H, Zhu D, Zhang G, Luo X, Xie W. AFAP1-AS1 promotes proliferation of pituitary adenoma cells through miR-103a-3p to activate PI3K/AKT signaling pathway. World Neurosurg. 2019;130(10):888–898. doi:10.1016/j.wneu.2019.07.032
33. Dasari C, Yaghnam DP, Walther R, Ummanni R. Tumor protein D52 (isoform 3) contributes to prostate cancer cell growth via targeting nuclear factor-kappaB transactivation in LNCaP cells. Tumour Biol. 2017;39(5):1010428317698382. doi:10.1177/1010428317698382
34. Li J, Li Y, Liu H, Liu Y, Cui B. The four-transmembrane protein MAL2 and tumor protein D52 (TPD52) are highly expressed in colorectal cancer and correlated with poor prognosis. PLoS One. 2017;12(5):e0178515. doi:10.1371/journal.pone.0178515
35. Kumamoto T, Seki N, Mataki H, et al. Regulation of TPD52 by antitumor microRNA-218 suppresses cancer cell migration and invasion in lung squamous cell carcinoma. Int J Oncol. 2016;49(5):1870–1880. doi:10.3892/ijo.2016.3690
36. Wang Y, Chen CL, Pan QZ, et al. Decreased TPD52 expression is associated with poor prognosis in primary hepatocellular carcinoma. Oncotarget. 2016;7(5):6323–6334. doi:10.18632/oncotarget.6319
37. Zhao Z, Liu H, Hou J, et al. Tumor protein D52 (TPD52) inhibits growth and metastasis in renal cell carcinoma cells through the PI3K/Akt signaling pathway. Oncol Res. 2017;25(5):773–779. doi:10.3727/096504016X14774889687280
38. Han G, Fan M, Zhang X. microRNA-218 inhibits prostate cancer cell growth and promotes apoptosis by repressing TPD52 expression. Biochem Biophys Res Commun. 2015;456(3):804–809. doi:10.1016/j.bbrc.2014.12.026
39. Zhang Y, Li Y, Wang J, Lei P. Long non‑coding RNA ferritin heavy polypeptide 1 pseudogene 3 controls glioma cell proliferation and apoptosis via regulation of the microRNA-224-5p/tumor protein D52 axis. Mol Med Rep. 2018;18(5):4239–4246. doi:10.3892/mmr.2018.9491
 © 2020 The Author(s). This work is published and licensed by Dove Medical Press Limited. The full terms of this license are available at https://www.dovepress.com/terms.php and incorporate the Creative Commons Attribution - Non Commercial (unported, v3.0) License.
By accessing the work you hereby accept the Terms. Non-commercial uses of the work are permitted without any further permission from Dove Medical Press Limited, provided the work is properly attributed. For permission for commercial use of this work, please see paragraphs 4.2 and 5 of our Terms.
© 2020 The Author(s). This work is published and licensed by Dove Medical Press Limited. The full terms of this license are available at https://www.dovepress.com/terms.php and incorporate the Creative Commons Attribution - Non Commercial (unported, v3.0) License.
By accessing the work you hereby accept the Terms. Non-commercial uses of the work are permitted without any further permission from Dove Medical Press Limited, provided the work is properly attributed. For permission for commercial use of this work, please see paragraphs 4.2 and 5 of our Terms.