Back to Journals » OncoTargets and Therapy » Volume 10
Long noncoding RNA CRNDE functions as a competing endogenous RNA to promote metastasis and oxaliplatin resistance by sponging miR-136 in colorectal cancer
Authors Gao H, Song X, Kang T, Yan B, Feng L, Gao L, Ai L, Liu X, Yu J, Li H
Received 1 July 2016
Accepted for publication 20 September 2016
Published 5 January 2017 Volume 2017:10 Pages 205—216
DOI https://doi.org/10.2147/OTT.S116178
Checked for plagiarism Yes
Review by Single anonymous peer review
Peer reviewer comments 3
Editor who approved publication: Professor Min Li
Hongyan Gao,1 Xiaodi Song,2 Ting Kang,3 Baohong Yan,4 Li Feng,5 Li Gao,6 Liang Ai,6 Xiaoni Liu,7,* Jie Yu,8,* Huiqi Li9,*
1Department of Gastroenterology, The First Affiliated Hospital of Xi’an Medical University, 2Department of Pharmacy, Xi’an Central Hospital, Xi’an, 3Department of Oncology, Yan’an University Affiliated Hospital, Yan’an, 4Department of Pharmacy, Hong-Hui Hospital, Xi’an Jiaotong University Medical College, Xi’an, 5Department of Anorectal Surgery, Ankang City Central Hospital, Ankang, 6Department of Pharmacy, Yan’an University Affiliated Hospital, 7Department of Endocrinology, Yan’an People’s Hospital, Yan’an, 8Department of General Surgery, Nuclear Industry 215 Hospital of Shaanxi Province, Xianyang, 9Department of General Surgery, The People’s Hospital of Baoji City, Baoji, People’s Republic of China
*These authors contributed equally to this work
Abstract: Colorectal neoplasia differentially expressed (CRNDE) is a novel gene recognized as a long noncoding RNA (lncRNA) that is highly elevated in colorectal cancer and many other solid tumors but its functions on metastasis and oxaliplatin (OXA) resistance are unknown. In our study, we confirmed the upregulation of CRNDE in both primary specimens from colorectal cancer patients and colorectal cancer cell lines. Knockdown of CRNDE expression inhibited the migration and invasion potency of colorectal cancer cells with no effect on cell apoptosis. Overexpression of CRNDE promoted the migration and invasion potency of colorectal cancer cells. Furthermore, we found that CRNDE conferred chemoresistance in colorectal cancer cells. Knockdown of CRNDE with OXA treatment decreased cell viability and promoted DNA damage and cell apoptosis, while the overexpression of CRNDE with OXA treatment reduced DNA damage and cell apoptosis. Further in-depth mechanistic studies revealed that CRNDE functioned as a competing endogenous RNA for miR-136, led to the de-repression of its endogenous target, E2F transcription factor 1 (E2F1). Overall, our findings demonstrate that CRNDE functions as a competing endogenous RNA to promote metastasis and OXA resistance by sponging miR-136 in colorectal cancer.
Keywords: CRNDE, colorectal cancer, metastasis, oxaliplatin resistance, miR-136, E2F1
Introduction
Colorectal cancer is still one of the most common cancers in the world, with high metastasis and recurrence rate being the most critical concerns.1,2 Although encouraging progress in diagnosis and cancer therapy has been achieved in the past decade, the 5-year overall survival rate is less than 10% in advanced disease and chemotherapy treatment remains essential for these patients. The activation of survival signaling pathways such as EGFR, PI3K/Akt, MAPK and STAT3 plays key roles in colon cancer initiation, development, progression and drug resistance, and therapies targeting those signaling axes are beneficial.3–6 The chemotherapy drugs such as 5-fluorouracil, oxaliplatin (OXA), irinotecan, cetuximab and bevacizumab are the first-line options for treatment of metastatic colorectal cancer.7–9 Increasing data have implicated the molecular mechanisms underlying resistance to OXA. The understanding of the molecular mechanisms underlying resistance to OXA is important to improve prediction of treatment response and guide treatment decisions in patients with metastatic colorectal cancer.
The novel gene colorectal neoplasia differentially expressed (CRNDE) transcripts are recognized as long noncoding RNAs (lncRNA) and located on chromosome 16 of the human genome. The expression of CRNDE is highly elevated in colorectal adenomas and carcinomas. CRNDE is the gene symbol or colorectal neoplasia differentially expressed, a novel gene the expression of which is highly elevated in colorectal adenomas and carcinomas (data from UCSC Genome Browser).10 Growing evidence has indicated that CRNDE acts through epigenetic mechanisms to regulate cell differentiation/pluripotency, which may relate to its deregulation in cancer. CRNDE expression is significantly upregulated in many cancers, including colorectal cancer and gliomas. Elevation in CRNDE expression has been shown to promote cell proliferation, migration and invasion while inhibiting apoptosis of glioma cells.10 Collective evidence has indicated that CRNDE is associated with adverse clinical characteristics and poor prognosis by regulating miRNAs in many solid tumors.11,12 However, the role of CRNDE in metastasis and OXA resistance of colorectal cancer and the depth mechanism are largely unknown.
In this study, we confirmed the upregulation of CRNDE in both primary specimens from colorectal cancer patients and colorectal cancer cell lines. CRNDE was knocked down by CRNDE siRNAs, and cell viability, migration and invasion potency of colorectal cancer cells were assessed. Our results showed that the knockdown of CRNDE inhibited the migration and invasion potency of colorectal cancer cells with no effect on cell apoptosis. We constructed plasmid pcDNA-CRNDE and pcDNA-Vector to ectopically express CRNDE and found that the overexpression of CRNDE promoted the migration and invasion potency of colorectal cancer cells. To determine whether CRNDE confers chemoresistance in colorectal cancer cells, the CRNDE knockdown and overexpressed HCT116 cells were treated with OXA. We found that the knockdown of CRNDE with OXA treatment decreased cell viability and promoted DNA damage and cell apoptosis, while the overexpression of CRNDE with OXA treatment reduced DNA damage and cell apoptosis.
Accumulated evidence showed that a range of lncRNA-sharing miRNA response elements (MREs) may act as a decoy to sequester miRNAs to prevent them from binding to targets and hence modulate many downstream target gene through translation. To gain insight into the possible mechanism, we used the bioinformatics databases to predict the potential lncRNA–miRNA interactions and found that miR-136 is a putative CRNDE-binding miRNA.
Previous studies reported that miR-136 plays a key role in temozolomide resistance by targeting the AEG-1 protein in glioma cell lines and miR-136 modulates the tumor sensitivity response to cisplatin by targeting E2F transcription factor 1 (E2F1) in glioma cells.13,14 In addition, the level of CRNDE and miR-136 in colorectal cancer tissues is inversely correlated by linear regression analysis. RNA immunoprecipitation assay and luciferase activity assays confirmed that CRNDE is the target of miR-136. We confirmed that E2F1 is a target of miR-136 in colorectal cancer cells and found that both gene and protein levels of E2F1 increased significantly in the CRNDE-overexpressed HCT116 cells. These observations indicate that CRNDE is a target of miR-136 and modulates the expression of E2F1.
Materials and methods
Cell culture and treatment
Human colorectal adenocarcinoma cell lines (SW480, HCT116 and HT-29) were obtained from American Type Culture Collection (Manassas, VA, USA) and were cultured in Dulbecco Modified Eagle Medium (DMEM) (Invitrogen, Life Technologies, Carlsbad, CA, USA) supplemented with 10% fetal bovine serum (FBS) (Gibco, Carlsbad, CA, USA). Human normal colon epithelial cell lines (HcoEpic and NCM460) were obtained from Xiehe Cell Bank of the Chinese Academy of Medical Sciences (Beijing, People’s Republic of China) and were cultured in McCoy’s 5A medium (Invitrogen, Life Technologies). Cells were maintained at 37°C in a water-saturated atmosphere with 5% CO2.
Clinical samples
The tumor tissues were collected from 10 colorectal cancer patients during surgery at Baoji City First People’s Hospital (Nanjing, People’s Republic of China). The study and the study protocol were approved by the institutional ethics committee of Baoji City First People’s Hospital. The tumor tissues were immediately frozen and kept at −80°C until assay. Written informed consent was obtained from all patients.
RNA isolation and quantitative real-time polymerase chain reaction
Total RNA was extracted from colorectal cancer tissues and cells using TRIzol reagent (Invitrogen). In all, 1 μg RNA was used as the template for single-strand cDNA synthesis utilizing random primers and the PrimeScript reverse transcriptase (M-MLV; Takara Bio, Shiga, Japan) according to the manufacturer’s instructions. Quantitative polymerase chain reaction (Q-PCR) was performed for CRNDE, GAPDH, E2F1, miR-136-5p and U6. The cDNA was amplified by an Applied Biosystems step-one plus sequence detection system (Applied Biosystems, Foster City, CA, USA). The relative expressions were normalized to endogenous controls using the comparative cycle threshold method, and fold change was calculated as 2−ΔΔCt in gene expression.
Cell viability assay
Cells were seeded in 96-well microtiter plates at a density of 3,000 cells/well. The cells were treated with OXA for 24 hours after transfected. The media was removed and fresh media was added to each well. Then, 10 μL of CCK-8 solution (CCK-8; Dojin, Kumamoto, Japan) was added into each well and incubated for 2 hours at 37°C. The absorbance was measured at 450 nm using the SpectraMax M5 microplate reader (Molecular Devices, Sunnyvale, CA, USA).
Apoptosis assay
Cell apoptosis was evaluated by annexin V-fluorescein isothiocyanate (FITC/PI) staining. Cells were collected and double stained with annexin V-FITC/PI according to the manufacturer’s instructions. Then, the cells were analyzed by an EPICS XL-MCL FAC scan (Becton-Dickinson, Mountain View, CA, USA) and the data were analyzed by CELL Quest 3.0 software (BD Biosciences, San Jose, CA, USA).
Cell migration assay
Wound healing assay was performed by measuring the movement of cells in a scraped, acellular area to assess cell motility. The wound closure was observed and photos were taken to assess the level of migration after 0 and 48 hours.
Matrigel invasion assay
Cell invasion through the Matrigel membrane was quantitated using Matrigel-coated transwell chambers (BD Biosciences) according to the manufacturer’s instructions. Cells (1×104) were seeded into the upper Matrigel-coated chambers with DMEM containing 10% FBS in the lower chamber. After 36 hours, non-invading cells in the upper chamber were removed by scrubbing with a cotton-tipped swab. Afterwards, the cells were fixed with 4% paraformaldehyde for 20 minutes and stained with 0.2% crystal violet (Sigma-Aldrich, St Louis, MO, USA) for 10 minutes. Six fields for each chamber were photographed using an inverted microscope and camera, and invading cells counted in each field.
Western blot analysis
Cells were homogenized in radio immunoprecipitation assay (RIPA) plus buffer in a buffer (50 mM Tris–Cl pH 8.0, 150 mM NaCl, 0.02% NaN3, 0.1% sodium dodecyl sulfate [SDS], 100 μg/mL phenylmethylsulfonyl fluoride, 1 μg/mL aprotinin, 1% Triton). After centrifugation, cell lysates (100 μg/lane) were subjected to 12% SDS-polyacrylamide gel electrophoresis and transferred onto polyvinylidene difluoride membranes (Millipore, Billerica, MA, USA). Antibodies against E2F1 (Santa Cruz, 1:1,000), GAPDH (CST, 1:1,000), γH2AX (CST, 1:2,000) and horseradish peroxidase-conjugated goat anti-rabbit secondary antibodies (Promab, 1:1,000) were used. Protein bands were detected by the enhanced chemiluminescence reaction, and blot film was scanned.
Cell transfection
siRNA oligonucleotides and negative control were designed from the ~350 base pair highly conserved gVC-In4 region within the CRNDE locus target sequence and synthesized by RiboBio Co Ltd (Guangzhou, People’s Republic of China). The pre-miR-136 (miRNA mimic), or its pre-control (scrambled negative controls), was designed and synthesized by RiboBio Co Ltd. The CRNDE full-length sequence was synthesized and subcloned into a pcDNA 3.1 vector (Invitrogen, Shanghai, People’s Republic of China). The cells were transfected with the aforementioned siRNA1, siRNA2, si-control, pre-miR-136, pre-control, pCDNA-CRNDE and vectors for 48 hours using Lipofectamine™ 3000 (Invitrogen, Shanghai, People’s Republic of China) following the manufacturer’s protocol. Cells were harvested after 48 hours for quantitative real-time polymerase chain reaction (qRT-PCR) to assess the efficiency of knockdown and overexpression.
RNA immunoprecipitation
The cells were rinsed with cold phosphate-buffered saline and lysed by a complete RNA lysis buffer with protease inhibitor and RNase inhibitor from an EZ-Magna RIP RNA-binding protein immunoprecipitation kit (Millipore) according to the manufacturer’s protocol. The cell lysates were stored at −80°C before use and supernatant from cell lysates was collected by high-speed centrifugation. The cell lysate was incubated with RIP immunoprecipitation buffer containing magnetic beads conjugated with human anti-Argonaute2 (Ago2) antibody (Abcam, Bristol, UK) and negative control normal mouse immunoglobulin G (IgG) (Sigma-Aldrich) overnight. The beads were rinsed with cold NT2 buffer (50 mM Tris-HCl, 150 mM NaCl, 1 mM MgCl2, 0.5% NP-40), followed by incubation with 10 mg/mL proteinase K buffer. The RNA bound to Ago2 antibody was extracted with TRIzol reagent. Then, the concentration and quality of RNA were measured. Furthermore, purified RNA was analyzed by qRT-PCR. RNA levels of CRNDE were presented as fold enrichment in Ago2 relative to IgG immunoprecipitates.
Luciferase reporter assay
Luciferase reporter plasmids (CRNDE-Wt and CRNDE-mut) were designed and constructed by Generay (Shanghai, People’s Republic of China). The HEK293T cells cultured in 24-well plates were co-transfected with luciferase reporter plasmids and miRNA mimics by Lipofectamine™ 3000 transfection reagent to analyze the interaction between CRNDE and miR-136. At 48 hours after transfection, luciferase activity assays were performed with the dual-luciferase reporter assay system.
Statistical analysis
All results are presented as mean ± standard error of mean of at least three independent experiments. Student’s t-test was used to assess differences between two groups, and one-way analysis of variance was used for multiple comparisons. A value of P<0.05 was considered to be statistically significant.
Results
CRNDE was increased in colorectal cancer tissues and cells
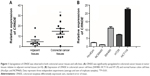
The relative expression levels of CRNDE were first assessed in colorectal cancer tissues and CRNDE expression was dramatically upregulated (Figure 1A). The expression levels of CRNDE in colorectal cancer cell lines and human normal colon cell lines were further measured. Notably, three cell lines (SW480, HCT116 and HT29) showed higher levels of CRNDE than normal cell lines (HcoEpic and NCM460) (Figure 1B). SW480 and HCT116 cell lines expressed relatively high CRNDE levels and they were selected for further study to assess the potential functional role of CRNDE.
Knockdown of CRNDE inhibited the migration and invasion of colorectal cancer cells
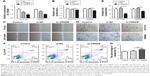
To investigate the biological role of CRNDE in colorectal cancer cells, the SW480 and HCT116 cells were transfected with CRNDE siRNAs or si-control to knockdown CRNDE in SW480 and HCT116 cells (Figure 2A). CCK8 assays showed that cell proliferation was decreased in SW480 and HCT116 cells transfected with CRNDE siRNA2 after 48 hours (Figure 2B). Wound healing assay revealed that cell migration potency of HCT116 and SW480 cells was inhibited by CRNDE knockdown (Figures 2C and S1A). Matrigel invasion assay revealed that the invasion potency of SW480 and HCT116 cells was also inhibited by CRNDE knockdown (Figure 2D and E). Cell apoptosis was evaluated and no significant change of cell apoptosis was found by CRNDE knockdown (Figure 2F and G).
Overexpression of CRNDE enhanced the migration and invasion of colorectal cancer cells
To further ascertain the role of CRNDE in proliferation, migration, invasion and apoptosis of colorectal cancer cells, we constructed plasmid pcDNA-CRNDE and pcDNA-Vector. The ectopic-expressed CRNDE in SW480 cells increased cell viability compared with transfection with pcDNA-Vector. However, there was no difference in HCT116 cells (Figure 3A). Wound healing assay revealed that cell migration potency of HCT116 and SW480 cells was enhanced by CRNDE overexpression (Figure 3B and Figure S1B). CRNDE overexpression enhanced the invasion potency of SW480 and HCT116 cells (Figure 3C and D). Furthermore, there is no significant change of cell apoptosis in CRNDE-overexpressed cells (Figure 3E).
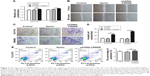
Knockdown of CRNDE enhances colorectal cancer cell sensitivity to OXA
To determine whether CRNDE confers chemoresistance in colorectal cancer cells, we transfected HCT116 cells with CRNDE siRNA or si-control and treated with OXA. As shown in Figure 4A, the knockdown of CRNDE alone had little effect on cell proliferation than control. However, the knockdown of CRNDE with OXA treatment had a strong effect on cell proliferation than control with OXA treatment. We also found the OXA drug resistance of HCT116 cells conferred by CRNDE was dose dependent (Figure 4B). OXA causes DNA crosslinking and stimulates H2AX phosphorylation at serine 139 to generate gH2AX as a major marker for DNA damage signaling in response to DNA double-strand break.15 OXA-triggered gH2AX in HCT116 cells transfected with CRNDE siRNA was elevated significantly, suggesting that CRNDE may control OXA resistance in colorectal cancer cells at the DNA damage induction level. (Figure 4C and D). OXA could bind to double-stranded DNA and form DNA adducts, interfering with DNA replication and ultimately triggering apoptosis. Hence, we examined whether CRNDE is involved in OXA-induced chemoresistance by affecting apoptosis. As shown in Figure 4E and F, the knockdown of CRNDE with OXA treatment induced the increased proportion of apoptosis cells. We have shown that the knockdown of CRNDE alone in HCT116 cells has no effect on apoptosis (Figure 2F). The results provided evidence that CRNDE enhances chemoresistance in colorectal cancer cells.
Overexpression of CRNDE enhances OXA chemoresistance of colorectal cancer cells
In order to further investigate the role of CRNDE in OXA resistance of colorectal cancer cells, the HCT116 cells were transfected with pcDNA-CRNDE or pcDNA-Vector. 48 hours later, cells were treated with OXA for 24 hours or 48 hours. The CRNDE ectopic-expressed HCT116 cells were treated with OXA, and reduction of cell viability was less evident compared with cells transfected with pcDNA-Vector (Figure 5A). The level of a major marker for DNA damage, gH2AX, was decreased when CRNDE ectopic-expressed HCT116 cells were treated with OXA compared with the control cells (Figure 5B). In addition, overexpression of CRNDE decreased the cell apoptosis proportion of HCT116 cells under the OXA treatment (Figure 5C and D). No impact of overexpression of CRNDE without OXA treatment on cell apoptosis was demonstrated (Figure 3E). Together, our findings demonstrated that CRNDE enhances colorectal cancer cell resistance to OXA.
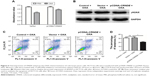
CRNDE is a target of miR-136 and modulates its target E2F1
Further in-depth study is required for a better understanding of the role of CRNDE in colorectal cancer cell metastasis and OXA resistance. Analysis with bioinformatics databases (Starbase, RNAhybrid) that predict potential lncRNA–miRNA interactions revealed that miR-136 is a putative CRNDE-binding miRNA (Figure 6A). We calculated the miR-136 levels in the colorectal cancer tissues and found that CRNDE and miR-136 level in the colorectal cancer tissue is inversely correlated by linear regression analysis (Figure 6B).
We supposed that CRNDE might harbor one miR-136 binding site and hijacking miR-136 by serving as a miRNA sponge. It has been shown that miRNA exerts its function by binding to Ago2, a core component of the RNA-induced silencing complex regulating miRNA-related gene expression changes. To assess our prediction, RNA-binding protein immunoprecipitation assay was performed. The results of RNA immunoprecipitation assay showed that CRNDE was preferentially enriched in Ago2-containing beads compared to the beads harboring control IgG antibody (Figure 6C). To further quantify our prediction that CRNDE could harbor one miR-136 binding site, wild-type and mutant CRNDE sequence containing the putative miR-136 recognition site was cloned downstream of the luciferase gene and co-transfected into HEK293T cells with pre-miR-136 or pre-control. Luciferase activity assays showed that miR-136 suppressed the activity of luciferase reporter harboring wild-type CRNDE but not the mutant CRNDE (Figure 6D). Collective data suggest that CRNDE acts as a miRNA decoy for miR-136.
It has been reported that E2F1 is a direct target of miR-136 in glioma cells.13 We performed a bioinformatics analysis by TargetScan algorithm (www.targetscan.org) and identified E2F1 as a predicted target of miR-136 (Figure 6E). Both gene and protein levels of E2F1 decreased in the HCT116 cells transfected by pre-miR-136 relative to pre-control (Figure 6F and G). We confirmed that E2F1 was a target of miR-136 in colorectal cancer cells. To determine whether CRNDE affects E2F1 expression, the HCT116 cells were transfected with pcDNA-Vector, pCDNA-CRNDE or pCDNA-CRNDE-mut (miR-136 and CRNDE binding sites were mutated) plasmid and gene and protein levels of E2F1 were tested. As shown in Figure 6H and I, both gene and protein levels of E2F1 increased significantly in CRNDE-overexpressed HCT116 cells compared with vector group. However, no change was found in HCT116 cells transfected with pCDNA-CRNDE-mut compared with vector plasmid.
Discussion
OXA is a platinum analog and the third-generation platinum drug used as adjuvant or first-line drug for treatment of metastatic colorectal cancer.16 OXA induces cytotoxicity mainly through the formation of platinum-DNA adducts resulting in DNA transcription and replication blockade and induces DNA damage-mediated cell apoptosis. OXA activates DNA damage repair and/or cell death signaling pathways.17 The molecular mechanisms of OXA chemoresistance are complex and multifactorial processes, such as drug influx/efflux modifications, DNA damage repair alterations, decrease of cell apoptosis and autophagy.18,19 The activation of survival signaling pathways is a major mechanism for OXA resistance. Previous studies reported that the expression of EGFR increased proportionally in relation to the level of acquired resistance to OXA in the tumor cell lines that are resistant to OXA.20 PI3K/Akt pathway activation was involved in OXA chemoresistance in human colon cancer.21,22
LncRNAs, more than 200 nucleotides in length, have emerged as critical regulators of human disease and prognostic markers in several cancers. LncRNAs are involved in tumor proliferation, metastasis and multiple cellular processes such as proliferation, migration, invasion, apoptosis and chemoresistance.16,18,23–26 Growing evidence has documented that miRNAs have important regulatory functions in biological processes that represent the hallmarks of cancer, such as proliferation, apoptosis, invasion and metastasis by targeting gene for deregulation or translational repression. What is more, collective data suggest that miRNAs are closely associated with the acquired chemoresistance in human carcinoma. In vitro, overexpression of miR-153, -203 and -143 has been associated with acquired resistance to OXA through modulation of FOXO3a, ATM kinase and IGF-1R, respectively.27–29 We predicted the potential lncRNA–miRNA interactions by bioinformatics databases and found that miR-136 is a putative CRNDE binding miRNA. The CRNDE-miR-136 interactions were confirmed by RNA immunoprecipitation assay and luciferase activity assays. E2F1 has been reported to be a direct target of miR-136 and involved in sensitization to chemotherapy in glioma cells.
E2F1 is a family member of eight (E2F1-8) transcription factors that regulate the cell cycle, migration, apoptosis and chemoresistance.30–32 The E2F1 expression was inhibited by pRb and released from phosphorylated pRB complexes leads to recruits coactivators to E2F-responsive genes in G1 phase transcription, DNA synthesis and DNA repair. E2F1 is aberrantly expressed in many malignancies, including melanoma, bladder tumor, lung tumor, prostate tumor, colorectal cancer and cisplatin-resistant ovarian cancer cell lines.33–38 E2F1 overexpression may promote proliferation or regulate cell apoptosis.39,40 It has been shown that miR-302b could enhance breast cancer cell sensitivity to cisplatin by targeting E2F1 and ATM. By this way, the cellular DNA damage response was enhanced.32 We confirmed that E2F1 is a target of miR-136 in colorectal cancer cells. To determine whether CRNDE affects E2F1 expression, we assessed the gene and protein levels of E2F1 in CRNDE-overexpressed HCT116 cells. Our results showed that both gene and protein levels of E2F1 increased significantly in the CRNDE-overexpressed HCT116 cells. Together, the collecting data indicate that CRNDE is a target of miR-136 and modulates the expression of E2F1.
CRNDE promotes metastasis and OXA resistance of colorectal cancer by functioning as a miR-136 sponge and modulating the expression of E2F1. Thus, our results shed light on utilizing CRNDE as a potential novel therapeutic target for the treatment of colorectal cancer.
Conclusion
In conclusion, CRNDE is involved in the migration and invasion of colorectal cancer cells. Furthermore, CRNDE conferred chemoresistance in colorectal cancer cells. Overexpression of CRNDE with OXA treatment reduced DNA damage and cell apoptosis. Further in-depth mechanistic studies revealed that CRNDE functions as a competing endogenous RNA to promote metastasis and OXA resistance by sponging miR-136 in colorectal cancer.
Acknowledgments
We are grateful to Shengjun Zhang, Xiaoli Guo, Hua Fan and Jiang Huang for curating clinical patient data and collecting patient samples. We are also grateful to Yaning Mu, Kang Zheng and Xiaodong Sun for expert technical assistance.
Disclosure
The authors report no conflicts of interest in this work.
References
Parkin DM, Bray F, Ferlay J, Pisani P. Global cancer statistics, 2002. CA Cancer J Clin. 2005;55(2):74–108. | ||
Gupta GP, Massagué J. Cancer metastasis: building a framework. Cell. 2006;127(4):679–695. | ||
Concha-Benavente F, Srivastava RM, Trivedi S, et al. Identification of the cell-intrinsic and -extrinsic pathways downstream of EGFR and IFNγ that induce PD-L1 expression in head and neck cancer. Cancer Res. 2016;76(5):1031–1043. | ||
Chen J. Signaling pathways in HPV-associated cancers and therapeutic implications. Rev Med Virol. 2015;25(suppl 1):24–53. | ||
Yu H, Lee H, Herrmann A, Buettner R, Jove R. Revisiting STAT3 signalling in cancer: new and unexpected biological functions. Nat Rev Cancer. 2014;14(11):736–746. | ||
Chen J, Zhang XD, Proud C. Dissecting the signaling pathways that mediate cancer in PTEN and LKB1 double-knockout mice. Sci Signal. 2015;8(392):pe1. | ||
Van Cutsem E, Rivera F, Berry S, et al. Safety and efficacy of first-line bevacizumab with FOLFOX, XELOX, FOLFIRI and fluoropyrimidines in metastatic colorectal cancer: the BEAT study. Ann Oncol. 2009;20(11):1842–1847. | ||
Redmond KL, Papafili A, Lawler M, Van Schaeybroeck S. Overcoming resistance to targeted therapies in cancer. Semin Oncol. 2015;42(6):896–908. | ||
Temraz S, Mukherji D, Alameddine R, Shamseddine A. Methods of overcoming treatment resistance in colorectal cancer. Crit Rev Oncol Hematol. 2014;89(2):217–230. | ||
Zheng J, Liu X, Wang P, et al. CRNDE promotes malignant progression of glioma by attenuating miR-384/PIWIL4/STAT3 axis. Mol Ther. 2016;24(7):1199–1215. | ||
Zheng J, Li XD, Wang P, et al. CRNDE affects the malignant biological characteristics of human glioma stem cells by negatively regulating miR-186. Oncotarget. 2015;6(28):25339–25355. | ||
Liu T, Zhang X, Yang YM, Du LT, Wang CX. Increased expression of the long noncoding RNA CRNDE-h indicates a poor prognosis in colorectal cancer, and is positively correlated with IRX5 mRNA expression. Onco Targets Ther. 2016;9:1437–1448. | ||
Chen W, Yang Y, Chen B, et al. MiR-136 targets E2F1 to reverse cisplatin chemosensitivity in glioma cells. J Neurooncol. 2014;120(1):43–53. | ||
Wu H, Liu Q, Cai T, Chen YD, Liao F, Wang ZF. MiR-136 modulates glioma cell sensitivity to temozolomide by targeting astrocyte elevated gene-1. Diagn Pathol. 2014;9:173. | ||
Revet I, Feeney L, Bruguera S, et al. Functional relevance of the histone gammaH2Ax in the response to DNA damaging agents. Proc Natl Acad Sci U S A. 2011;108(21):8663–8667. | ||
Schmitt AM, Chang HY. Gene regulation: long RNAs wire up cancer growth. Nature. 2013;500(7464):536–537. | ||
Kelland L. The resurgence of platinum-based cancer chemotherapy. Nat Rev Cancer. 2007;7(8):573–584. | ||
Holohan C, Van Schaeybroeck S, Longley DB, Johnston PG. Cancer drug resistance: an evolving paradigm. Nat Rev Cancer. 2013;13(10):714–726. | ||
Martinez-Balibrea E, Martínez-Cardús A, Ginés A, et al. Tumor-related molecular mechanisms of oxaliplatin resistance. Mol Cancer Ther. 2015;14(8):1767–1776. | ||
Ekblad L, Johnsson A. Cetuximab sensitivity associated with oxaliplatin resistance in colorectal cancer. Anticancer Res. 2012;32(3):783–786. | ||
Chen J, Huang XF, Qiao L, Katsifis A. Insulin caused drug resistance to oxaliplatin in colon cancer cell line HT29. J Gastrointest Oncol. 2011;2(1):27–33. | ||
Nemoto S, Nakamura M, Osawa Y, et al. Sphingosine kinase isoforms regulate oxaliplatin sensitivity of human colon cancer cells through ceramide accumulation and Akt activation. J Biol Chem. 2009;284(16):10422–10432. | ||
Bartonicek N, Maag JL, Dinger ME. Long noncoding RNAs in cancer: mechanisms of action and technological advancements. Mol Cancer. 2016;15(1):43. | ||
Lavorgna G, Vago R, Sarmini M, Montorsi F, Salonia A, Bellone M. Long non-coding RNAs as novel therapeutic targets in cancer. Pharmacol Res. 2016;110:131–138. | ||
Riquelme I, Ili C, Roa JC, Brebi P. Long non-coding RNAs in gastric cancer: mechanisms and potential applications. Oncotarget. Epub 2016 May 16. | ||
Qi P, Zhou XY, Du X. Circulating long non-coding RNAs in cancer: current status and future perspectives. Mol Cancer. 2016;15(1):39. | ||
Qian X, Yu J, Yin Y, et al. MicroRNA-143 inhibits tumor growth and angiogenesis and sensitizes chemosensitivity to oxaliplatin in colorectal cancers. Cell Cycle. 2013;12(9):1385–1394. | ||
Zhang L, Pickard K, Jenei V, et al. miR-153 supports colorectal cancer progression via pleiotropic effects that enhance invasion and chemotherapeutic resistance. Cancer Res. 2013;73(21):6435–6447. | ||
Zhou Y, Wan G, Spizzo R, et al. miR-203 induces oxaliplatin resistance in colorectal cancer cells by negatively regulating ATM kinase. Mol Oncol. 2014;8(1):83–92. | ||
Crosby ME, Almasan A. Opposing roles of E2Fs in cell proliferation and death. Cancer Biol Ther. 2004;3(12):1208–1211. | ||
Ma Y, Xin Y, Li R, et al. TFDP3 was expressed in coordination with E2F1 to inhibit E2F1-mediated apoptosis in prostate cancer. Gene. 2014;537(2):253–259. | ||
Cataldo A, Cheung DG, Balsari A, et al. miR-302b enhances breast cancer cell sensitivity to cisplatin by regulating E2F1 and the cellular DNA damage response. Oncotarget. 2016;7(1):786–797. | ||
Chang WY, Andrews J, Carter DE, Dagnino L. Differentiation and injury-repair signals modulate the interaction of E2F and pRB proteins with novel target genes in keratinocytes. Cell Cycle. 2006;5(16):1872–1879. | ||
Li Z, Hou M, Wang H, Wang Z. A randomized study of gemcitabine plus oxaliplatin versus gemcitabine plus cisplatin as the 1st line chemotherapy for advanced non-small cell lung cancer in elderly patients. Zhongguo Fei Ai Za Zhi. 2011;14(7):588–592. | ||
Laine A, Westermarck J. Molecular pathways: harnessing E2F1 regulation for prosenescence therapy in p53-defective cancer cells. Clin Cancer Res. 2014;20(14):3644–3650. | ||
Knoll S, Emmrich S, Putzer BM. The E2F1-miRNA cancer progression network. Adv Exp Med Biol. 2013;774:135–147. | ||
Vélez-Cruz R, Johnson DG. E2F1 and p53 transcription factors as accessory factors for nucleotide excision repair. Int J Mol Sci. 2012;13(10):13554–13568. | ||
Engelmann D, Pützer BM. The dark side of E2F1: in transit beyond apoptosis. Cancer Res. 2012;72(3):571–575. | ||
Zhan L, Zhang Y, Wang W, Song E, Fan Y, Wei B. E2F1: a promising regulator in ovarian carcinoma. Tumour Biol. 2016;37(3):2823–2831. | ||
Poppy Roworth A, Ghari F, La Thangue NB. To live or let die – complexity within the E2F1 pathway. Mol Cell Oncol. 2015;2(1):e970480. |
Supplementary material
 © 2017 The Author(s). This work is published and licensed by Dove Medical Press Limited. The full terms of this license are available at https://www.dovepress.com/terms.php and incorporate the Creative Commons Attribution - Non Commercial (unported, v3.0) License.
By accessing the work you hereby accept the Terms. Non-commercial uses of the work are permitted without any further permission from Dove Medical Press Limited, provided the work is properly attributed. For permission for commercial use of this work, please see paragraphs 4.2 and 5 of our Terms.
© 2017 The Author(s). This work is published and licensed by Dove Medical Press Limited. The full terms of this license are available at https://www.dovepress.com/terms.php and incorporate the Creative Commons Attribution - Non Commercial (unported, v3.0) License.
By accessing the work you hereby accept the Terms. Non-commercial uses of the work are permitted without any further permission from Dove Medical Press Limited, provided the work is properly attributed. For permission for commercial use of this work, please see paragraphs 4.2 and 5 of our Terms.