Back to Journals » OncoTargets and Therapy » Volume 8
Long noncoding RNA CCAT2 promotes breast tumor growth by regulating the Wnt signaling pathway
Received 14 June 2015
Accepted for publication 26 July 2015
Published 21 September 2015 Volume 2015:8 Pages 2657—2664
DOI https://doi.org/10.2147/OTT.S90485
Checked for plagiarism Yes
Review by Single anonymous peer review
Peer reviewer comments 5
Editor who approved publication: Dr Faris Farassati
Video abstract presented by Dong Zhang
Views: 256
Yi Cai,1,* Jing He,2,* Dong Zhang1
1Department of Geriatric Oncology, 2Department of Geriatric Integrated Surgery, The General Hospital of Chinese People’s Liberation Army, Beijing City, People’s Republic of China
*These authors contributed equally to this work
Abstract: In addition to protein-coding genes, the human genome makes a large amount of noncoding RNAs, including microRNAs and long noncoding RNAs (lncRNAs). Emerging evidence indicates that lncRNAs could have a critical role in the regulation of cellular processes such as cell growth and apoptosis as well as cancer progression and metastasis. The lncRNA CCAT2 is dysregulated in several cancers such as colon cancer, non-small cell lung cancer, esophageal squamous cell carcinoma, gastric cancer, and breast cancer; however, the contributions of CCAT2 to breast cancer remain largely unknown. In the current paper, we first confirmed the high expression level of CCAT2 in breast cancer tissues and breast cancer cell lines by reverse transcription quantitative polymerase chain reaction (RT-qPCR) assay, and we further analyzed the relationship between CCAT2 expression and clinical prognostic factors. Also, the biological function of CCAT2 was explored and the results showed silencing of CCAT2 could suppress cell growth in vitro and tumor formation in vivo. Finally, our results revealed that the abnormal expression of CCAT2 could influence the Wnt signaling pathway. In conclusion, lncRNA CCAT2 might be considered as a novel molecule involved in breast cancer development, which provides a potential therapeutic target for breast cancer.
Keywords: long noncoding RNAs, CCAT2, breast cancer, Wnt signaling pathway
Introduction
Globally, breast cancer is the most commonly diagnosed cancer in women, while metastatic disease is the leading cause of cancer-related deaths in this group.1 Therapeutic failure and distant metastasis have been a major challenge in the treatment of breast cancer. Thus, exploring more markers to predict responsiveness of treatment, tumor progression, and potential target therapies is becoming more and more important.2,3
As we all know, cancer progression involves various changes in gene expression. As a newly discovered class of noncoding genes, emerging evidence suggests that long noncoding RNAs (lncRNAs) may serve as master gene regulators capable of controlling protein-coding and noncoding genes.4 The altered expression of lncRNAs is frequently observed in human cancers, including breast cancer. Alterations in the expression, the primary structure, and secondary structure of lncRNAs as well as changes in their binding proteins are often associated with metastasis, invasion, and patient survival.5 These findings suggested that the potential role of lncRNAs should be further investigated.
Ling et al first discovered CCAT2, a novel noncoding RNA mapping to 8q24, in 2013. They found that CCAT2 is highly overexpressed in microsatellite-stable colorectal cancer and promotes tumor growth, metastasis, and chromosomal instability.6 Redis et al showed that CCAT2 may represent a valuable predictive marker of clinical outcomes (shorter metastases-free survival and overall survival) for a specific subgroup of breast cancer patients.7 Qiu et al reported that CCAT2 is a lung adenocarcinoma-specific lncRNA and promotes invasion of non-small cell lung cancer, and CCAT2 could be used as a potential biomarker for lymph node metastasis.8 Wang et al reported that CCAT2 correlates with smoking in esophageal squamous cell carcinoma, and that CCAT2 showed higher diagnostic performance than conventional serum biomarkers.9 In addition, Wang et al reported that upregulation of CCAT2 was correlated with gastric cancer progression, and that CCAT2 might be a potential molecular biomarker for predicting the prognosis of patients.10 These authors suggest that CCAT2 might exert its biological function as an oncogene in several kinds of cancers; however, the detailed function of CCAT2 in breast cancer was still uncertain.
In the current paper, we investigated the expression of CCAT2 in breast cancer tissues and cell lines. We then examined the relationships between CCAT2 expression levels in tumor tissues and the clinical prognostic features of breast cancer patients. Furthermore, we conducted in vitro assays to demonstrate the biological functions of CCAT2 in the proliferation of breast cancer cells. Finally, the in vivo assay of the tumor xenograft model was carried out to verify the ability of tumor formation.
Materials and methods
Patients’ samples
The Ethical Committee of The General Hospital of Chinese People’s Liberation Army for Clinical Research approved this study. Paired breast cancer tissue and adjacent normal breast tissue were obtained from 67 patients who had undergone surgical breast cancer resection between March 2009 and February 2010 at the Department of Geriatric Oncology and the Department of Geriatric Comprehensive Surgery, in the General Hospital of Chinese People’s Liberation Army. All the participators are female. Average age of breast cancer patients was 51.5±12.5 years (range: 29–71), and the body mass index, considered as a risk factor for breast cancer, was 22.4±2.5 kg/m2 (range: 19–34). No smoking subjects were included in our study. All the patients were diagnosed by two experienced pathologists according to American Society of Clinical Oncology breast cancer guidelines. The non-tumorous tissue samples were at least 2 cm from the edge of the tumor, contained no obvious tumor cells, and were also evaluated by the pathologists. The tissue samples were obtained from fresh surgical specimens, then frozen in liquid nitrogen immediately, and stored at −80°C. All the samples were obtained with the patients’ informed consent.
Cell lines and culture conditions
The human breast cancer cell lines MDA-MB-231 and MCF-7 were purchased from Beijing Zhongyuan Ltd. (Beijing, People’s Republic of China). Normal mammary fibroblast cell line Hs578Bst was purchased from Shanghai Kexing Biotech Ltd. (Shanghai, People’s Republic of China). MDA-MB-231 cells were cultured in L-15 medium, and MCF-7 cells and Hs578Bst cells were cultured in Dulbecco’s Modified Eagle’s Medium. All media included 10% fetal bovine serum, 100 U/mL of penicillin, and 100 μg/mL of streptomycin. Both cell lines were cultured at 37°C and 5% CO2.
Cell transfection
The siRNA specifically targeting CCAT2 (si-CCAT2) was commercially constructed by Shanghai GenePharma Co. Ltd (Shanghai, People’s Republic of China), the sequences were S: 5′-GUGCAACUCUGCAAUUUAAUU-3′, S: 5′-UUAAAUUGCAGAGUUGCACUU-3′, and the scrambled nucleotide was used as the negative control (si-NS). Cells were transfected with siRNAs using Lipofectamine RNAiMAX reagent (Thermo Fisher Scientific, Waltham, MA, USA) according to the manufacturer’s protocol.
RNA extraction and quantitative reverse transcription polymerase chain reaction
RNA was isolated from tissues or cells using TRIzol reagent (Thermo Fisher Scientific), and then the RNA was reverse transcribed using SuperScript First Strand cDNA System (Thermo Fisher Scientific) according to the manufacturer’s instructions. RT-qPCR was performed to detect lncRNA expression, and delta Ct values were used to determine their absolute expression, delta-delta Ct values were used to determine their relative expression as fold changes occur. The PCR amplification was performed for 40 cycles at 94°C for 30 seconds, 60°C for 30 seconds, and 72°C for 30 seconds, on a Thermo Fisher Scientific 7300 Real-Time PCR System with 1.0 μL of cDNA and SYBR Green Real-time PCR Master Mix (TransGen Biotech, Beijing, People’s Republic of China). The primers used are listed below: CCAT2: S: 5′-AGACAGTGCCAGCCAACC-3′, AS: 5′-TGCCAAACCCTTCCCTTA-3′; glyceraldehyde-3-phosphate dehydrogenase (GAPDH) was used as an internal control, S: 5′-AATGGACAACTGGTCGTGGAC-3′, and AS: 5′-CCCTCCAGGGGATCTGTTTG-3′.
Western blot analysis
The protein samples were extracted from cultured cells and both the cytoplasmic and nuclear protein were separated and extracted using Nucleoprotein Extraction Kit (purchased from Sangon Biotech, Shanghai, People’s Republic of China) according to the manufacturer’s instructions. The protein content of each tube was calculated via bicinchoninic acid protein quantification method. The protein samples (40 μg) were fractionated by sodium dodecyl sulfate polyacrylamide gel electrophoresis (10% polyacrylamide gel). Mouse monoclonal primary antibodies against β-catenin and Actin (C-2) (Santa Cruz Biotechnology Inc., Dallas, TX, USA) and mouse monoclonal primary antibodies against histone 3 (Zhongshanjinqiao Biotech, Beijing, People’s Republic of China) were used for the Western blot.
Luciferase assay
The TOP/FOP Flash luciferase reporter plasmid system was purchased from Biovector NTCC Ltd (Beijing, People’s Republic of China). Luciferase assays were performed using a luciferase assay kit from Vigorous Biotech (Beijing, People’s Republic of China) according to the manufacturer’s protocol. First, the MCF-7 and MDA-MB-231 cells were transfected with the appropriate plasmids in 24-well plates. Then, the cells were harvested and lysed for a luciferase assay 24 hours after transfection. Firefly luciferase was used as a base line, and Renilla luciferase was used as the internal control.
MTT assay
The MCF-7 and MDA-MB-231 cells transfected with either siCCAT or siNC for 48 hours were suspended and seeded into 96-well plates with a cell density of 5×103/well, and the final volume was 150 μL/well. MTT solution (20 μL) was added to the plates 12, 24, 48, and 72 hours later. The cells were cultured for another 4 hours at 37°C. Then, the medium was discarded and 150 μL of dimethyl sulfoxide was added and the cells were oscillated for 15 minutes. The absorbance was measured at 490 nm using an enzyme-labeled analyzer.
Tumor xenograft model
Female nude mice (4–5 weeks old) were purchased from Vital River Laboratory Animal Technology Ltd. (Beijing, People’s Republic of China). MCF-7 cells were transfected with si-CCAT (100 nM) or siNC (100 nM) respectively. MCF-7 (2×106 cells) in 50% matrigel (BD Biosciences, San Jose, CA, USA) were subcutaneously injected into the right flank of nude mice to develop the xenograft mouse models bearing tumors. The tumor-cell injection was performed as described previously.11 For the tumor growth assay, tumor volume was calculated as follows: tumor volume=width2×length/2. The mice were sacrificed at day 24 and the tumors were separated for weighing. All experimental protocols were performed in accordance with ARRIVE (Animal Research: Reporting In Vivo Experiments) guidelines of the UK and were approved by the Institutional Animal Care and Use Committee of the General Hospital of Chinese People’s Liberation Army.
Statistical analysis
All statistical analyses were carried out using SPSS 17.0 (SPSS Inc., Chicago, IL, USA). Most of the data were analyzed using independent two-tailed Student’s t-test, and overall survival was estimated by using the Kaplan–Meier method. Values of P<0.05 were considered statistically significant.
Results
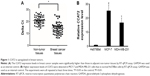
CCAT2 is upregulated in breast tumors
First, we measured the CCAT2 expression levels by RT-qPCR in a set of 67 matched samples. The result showed that CCAT2 appeared to have higher expression in breast cancer tissues than in adjacent non-tumor tissues (P<0.05) (Figure 1A). The CCAT2 expression was also examined by RT-qPCR in human breast cancer cell lines MDA-MB-231 and MCF-7. The normal mammary fibroblast cell line Hs578Bst was used as the control. This experiment showed that CCAT2 expression was higher in breast cancer cell lines than in normal mammary fibroblasts (P<0.001) (Figure 1B).
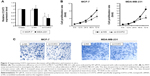
CCAT2 expression correlated with clinical prognostic factors
To assess the correlation between CCAT2 expression and prognostic factors, we divided the breast cancer patients into two groups according to the CCAT2 expression level in the tumor tissues: low CCAT2 expression group (the change of relative expression < eightfold) and high CCAT2 expression group (the change of relative expression >eightfold) (Figure 2A). As shown in Figure 2B, patients with high CCAT2 expression had a significantly poorer prognosis than those with low expression, and the relative level of CCAT2 expression was correlated with overall survival rate of patients with breast cancer (Figure 2B). Overall, these observations indicated that increased CCAT2 expression is associated with the progression and development of breast cancer.
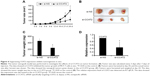
Suppressing CCAT2 expression decreases cell proliferation and invasion in vitro
To further investigate the role of CCAT2 in human breast cancer cells, si-CCAT2 was designed and transfected into MCF-7 and MDA-MB-231 breast cancer cells. Nonspecific siRNA (si-NS) was used as the negative control. As shown in Figure 3A, cells transfected with si-CCAT2 presented a significantly decreased mRNA expression level of CCAT2 compared with the si-NS group in both cells (P<0.05) (Figure 3A). The MTT assay showed that the proliferation rate was downregulated in the si-CCAT2-transfected breast cancer cells after 48 hours compared with the si-NS group (P<0.05) (Figure 3B). Furthermore, to analyze the role of CCAT2 in cell invasion, transwell assays were performed in MCF-7 and MDA-MB-231 cells, and the result indicated that the invasiveness of both the MCF-7 and MDA-MB-231 cells transfected with si-CCAT2 were greatly decreased compared with the si-NS groups, respectively (Figure 3C). All together, the data demonstrated that suppression of CCAT2 expression could inhibit cell proliferation and invasion of breast cancer cells in vitro.
Suppressing CCAT2 expression inhibits tumorigenesis in vivo
To further determine the role of CCAT2 on tumorigenesis, MCF-7 cells transfected with either si-NS or si-CCAT2 mixed with matrigel were injected into nude mice. Two weeks after injection, a palpable tumor could be observed, and the results were consistent with the in vitro study; si-CCAT2 significantly reduced tumor growth at the indicated time (Figure 4A). In addition, tumors derived from the si-CCAT2 group grew at a slower rate than the si-NS group (Figure 4B), and the tumor weight in the si-CCAT2 group was significantly less than the si-NS group (Figure 4C). The expression level of CCAT2 was also detected, and the result showed that CCAT2 was knocked down effectively (Figure 4D).
Suppressing CCAT2 expression affects the Wnt/β-catenin signaling pathway
To better understand the detailed regulation mechanism of CCAT2 in breast cancer, we first examined whether suppressing CCAT2 affects the Wnt signaling pathway, whose activation plays an important role in breast cancer development. Using the si-CCAT2 group and the si-NS group of the MCF-7 and MDA-MB-231 cells, as well as the Wnt signaling inhibitor FH 535, we found that suppressing the expression of CCAT2 decreased the levels of β-catenin both in the cytoplasm and nucleus via Western blot (Figure 5A), but FH 535 only affected the β-catenin recruitment, and the combination of si-CCAT2 and FH 535 could induce a synergetic effect on Wnt signaling activity. Moreover, CCAT2 knockdown reduced the expression of CCND1 and c-myc (classic downstream genes of the Wnt/β-catenin signaling pathway) via RT-qPCR in both MCF-7 and MDA-MB-231 cells (Figure 5B). In addition, the TOP/FOP Flash luciferase reporter system was employed in MCF-7 and MDA-MB-231 cells; the result showed that suppressing CCAT2 expression inhibited the Wnt/β-catenin signaling pathway transcriptional activity, and combination of siCCAT2 and FH 535 could synergistically inhibit the Wnt signaling (Figure 5C). Thus, these results suggest that suppressing CCAT2 expression affects the Wnt/β-catenin signaling pathway.
Discussion
Accumulating evidence shows that lncRNAs play more and more important roles in a wide range of biological processes. Aberrant lncRNA expression is involved in many cancers, and lncRNAs have also been found to function as new regulators in cancer development.12,13 For example, Huang et al reported that lncRNA ANRIL expression was elevated in hepatocellular carcinoma tissues, and the expression of ANRIL was significantly associated with tumor size and liver cancer stage. In addition, knockdown of ANRIL expression could impair cell proliferation and invasion and induce cell apoptosis both in vitro and in vivo.14 Wang et al presented that lncRNA MEG3 was greatly downregulated in papillary thyroid carcinoma tissues with lymph node metastasis compared with primary thyroid cancer, and that the downregulation of MEG3 correlated with lymph node metastasis. Biologically, overexpression of MEG3 could inhibit the cell migration and invasion in thyroid cancer cell lines.15 These authors have shown that lncRNAs can function as oncogenes or tumor suppressors during cancer progression.
Recently, identification of lncRNAs as biomarkers has helped us to diagnose cancer earlier. For example, Isin et al investigated the exosomal lncRNA GAS5 and lincRNA-p21 levels in urine samples from 30 patients with prostate cancer and 49 patients with benign prostatic hyperplasia. They observed a significant difference in the exosomal lncRNA-p21 levels between prostate cancer and benign prostatic hyperplasia patients.16 Xu et al evaluated the feasibility and clinical significance of circulating serum lncRNA RP11-445H22.4 as a biomarker for the detection of breast cancer, and they found a correlation between the levels of serum lncRNA RP11-445H22.4 in breast cancer patients and the clinicopathological factors of these patients.17 Identification of special lncRNAs as biomarkers may help us improve the diagnostic prediction of the malignant state of patients with cancer. Some investigators have begun to identify lncRNAs whose expression is associated with aberrant signaling or unregulated survival of breast cancer cells.18 The overexpression of lncRNA HOTAIR has been associated with enhanced metastasis and invasion of breast cancer cells and can be used as a predictor of overall survival and progression-free survival.19 The expression of lncRNA GAS5 is downregulated in breast cancer samples relative to adjacent unaffected normal breast tissue, and has a distinct tumor suppressive role in breast cancer by inducing apoptosis and suppressing cell proliferation.20 The expression of CCAT2 in normal breast tissue and breast cancer tissue was evaluated, and the results showed that CCAT2 may represent a valuable predictive marker of clinical outcomes (shorter metastases-free survival and overall survival) for a specific subgroup of breast cancer patients, in whom high levels of CCAT2 will indicate that these patients will not benefit from CMF (cyclophosphamide, methotrexate, and 5-fluorouracil) adjuvant chemotherapy.7 In our research, we presented that the expression of CCAT2 was significantly upregulated in breast cancer tissues and breast cancer cell lines, and the expression of CCAT2 correlated with clinical prognostic factors. Biologically, suppressing the expression of CCAT2 could inhibit cell proliferation and invasion in vitro and tumor formation in vivo. With the use of innovative technologies, it is now possible to find more ncRNAs used for diagnosis and prognosis of breast cancer. The biological functions and molecular mechanisms for these lncRNAs should be fully explored in the future.
Accumulating evidence indicates that the Wnt/β-catenin signaling pathway contributes to the neoplastic process, of which β-catenin is one of the key downstream effectors.21 An inappropriate activation of the Wnt/β-catenin signaling pathway leads to the development of human cancers, including breast cancer.22 The upstream factors of the Wnt/β-catenin signaling pathway and their regulation of carcinogenesis and metastasis in breast cancers is still not well understood.23 In the current paper, we found that the abnormal expression of CCAT2 could influence the Wnt signaling pathway by suppressing β-catenin activity. Using cytoplasmic and nuclear protein isolation, we found that knockdown of CCAT2 suppressed both the cytoplasmic and nuclear β-catenin level, suggesting that CCAT2 regulates β-catenin level at the transcriptional level; however, the Wnt signaling inhibitor FH 535, which only affects the β-catenin recruitment to target gene promoter, did not affect the cytoplasm β-catenin but only affected the nuclear accumulation of β-catenin. Nevertheless, combination of si-CCAT2 and FH 535 in breast cancer cells indeed had a synergetic effect on Wnt signaling activity. Given the fact that CCAT2 regulates Wnt signaling, it would be helpful for personalized therapy in breast cancer. For example, for patients with high CCAT2 expression, targeting Wnt signaling could be a therapeutic strategy.
Conclusion
Overall, our findings broaden the scope of known lncRNA CCAT2 functions and aids in the discovery and design of more effective and evidence-based therapeutic targets for the treatment of breast cancer.
Disclosure
The authors report conflict of interest in this work.
References
Pelkonen M, Luostari K, Tengstrom M, et al. Low expression levels of hepsin and TMPRSS3 are associated with poor breast cancer survival. BMC Cancer. 2015;15:431. | ||
Lv ZD, Kong B, Liu XP, et al. CXCL12 chemokine expression suppresses human breast cancer growth and metastasis in vitro and in vivo. Int J Clin Exp Pathol. 2014;7(10):6671–6678. | ||
Kabbage M, Trimeche M, Ben Nasr H, et al. Expression of the molecular chaperone alpha B-crystallin in infiltrating ductal breast carcinomas and the significance thereof: an immunohistochemical and proteomics-based strategy. Tumour Biol. 2012;33(6):2279–2288. | ||
Huang J, Zhou N, Watabe K, et al. Long non-coding RNA UCA1 promotes breast tumor growth by suppression of p27 (Kip1). Cell Death Dis. 2014;5:e1008. | ||
Chi Y, Huang S, Yuan L, et al. Role of BC040587 as a predictor of poor outcome in breast cancer. Cancer Cell Int. 2014;14(1):123. | ||
Ling H, Spizzo R, Atlasi Y, et al. CCAT2, a novel noncoding RNA mapping to 8q24, underlies metastatic progression and chromosomal instability in colon cancer. Genome Res. 2013;23(9):1446–1461. | ||
Redis RS, Sieuwerts AM, Look MP, et al. CCAT2, a novel long non-coding RNA in breast cancer: expression study and clinical correlations. Oncotarget. 2013;4(10):1748–1762. | ||
Qiu M, Xu Y, Yang X, et al. CCAT2 is a lung adenocarcinoma-specific long non-coding RNA and promotes invasion of non-small cell lung cancer. Tumour Biol. 2014;35(6):5375–5380. | ||
Wang J, Qiu M, Xu Y, et al. Long noncoding RNA CCAT2 correlates with smoking in esophageal squamous cell carcinoma. Tumour Biol. 2015;36(7):5523–5528. | ||
Wang CY, Hua L, Yao KH, Chen JT, Zhang JJ, Hu JH. Long non-coding RNA CCAT2 is up-regulated in gastric cancer and associated with poor prognosis. Int J Clin Exp Pathol. 2015;8(1):779–785. | ||
De P, Sun Y, Carlson JH, Friedman LS, Leyland-Jones BR, Dey N. Doubling down on the PI3K-AKT-mTOR pathway enhances the antitumor efficacy of PARP inhibitor in triple negative breast cancer model beyond BRCA-ness. Neoplasia. 2014;16(1):43–72. | ||
Gerasimova E, Audit B, Roux SG, et al. Wavelet-based multifractal analysis of dynamic infrared thermograms to assist in early breast cancer diagnosis. Front Physiol. 2014;5:176. | ||
Hauptman N, Glavac D. Long non-coding RNA in cancer. Int J Mol Sci. 2013;14(3):4655–4669. | ||
Huang M, Chen WM, Qi FZ, et al. Long non-coding RNA ANRIL is up-regulated in hepatocellular carcinoma and promotes cell apoptosis by epigenetically silencing of KLF2. J Hematol Oncol. 2015;8:50. | ||
Wang C, Yan G, Zhang Y, Jia X, Bo P. Long non-coding RNA MEG3 suppresses migration and invasion of thyroid carcinoma by targeting of Rac1. Neoplasma. 2015;62(4):541–549. | ||
Isin M, Uysaler E, Ozgur E, et al. Exosomal lncRNA-p21 levels may help to distinguish prostate cancer from benign disease. Front Genet. 2015;6:168. | ||
Xu N, Chen F, Wang F, et al. Clinical significance of high expression of circulating serum lncRNA RP11-445H22.4 in breast cancer patients: a Chinese population-based study. Tumour Biol. Epub 2015 May 1. | ||
Hayes EL, Lewis-Wambi JS. Mechanisms of endocrine resistance in breast cancer: an overview of the proposed roles of noncoding RNA. Breast Cancer Res. 2015;17:40. | ||
Gupta RA, Shah N, Wang KC, et al. Long non-coding RNA HOTAIR reprograms chromatin state to promote cancer metastasis. Nature. 2010;464(7291):1071–1076. | ||
Mourtada-Maarabouni M, Pickard MR, Hedge VL, Farzaneh F, Williams GT. GAS5, a non-protein-coding RNA, controls apoptosis and is downregulated in breast cancer. Oncogene. 2009;28(2):195–208. | ||
Nelson WJ, Nusse R. Convergence of Wnt, beta-catenin, and cadherin pathways. Science. 2004;303(5663):1483–1487. | ||
Hwang MS, Yu N, Stinson SY, et al. miR-221/222 targets adiponectin receptor 1 to promote the epithelial-to-mesenchymal transition in breast cancer. PLoS One. 2013;8(6):e66502. | ||
Cai K, Jiang L, Wang J, et al. Downregulation of beta-catenin decreases the tumorigenicity, but promotes epithelial-mesenchymal transition in breast cancer cells. J Cancer Res Ther. 2014;10(4):1063–1070. |
 © 2015 The Author(s). This work is published and licensed by Dove Medical Press Limited. The full terms of this license are available at https://www.dovepress.com/terms.php and incorporate the Creative Commons Attribution - Non Commercial (unported, v3.0) License.
By accessing the work you hereby accept the Terms. Non-commercial uses of the work are permitted without any further permission from Dove Medical Press Limited, provided the work is properly attributed. For permission for commercial use of this work, please see paragraphs 4.2 and 5 of our Terms.
© 2015 The Author(s). This work is published and licensed by Dove Medical Press Limited. The full terms of this license are available at https://www.dovepress.com/terms.php and incorporate the Creative Commons Attribution - Non Commercial (unported, v3.0) License.
By accessing the work you hereby accept the Terms. Non-commercial uses of the work are permitted without any further permission from Dove Medical Press Limited, provided the work is properly attributed. For permission for commercial use of this work, please see paragraphs 4.2 and 5 of our Terms.