Back to Journals » Cancer Management and Research » Volume 12
Long Noncoding MAGI2-AS3 Suppresses Several Cellular Processes of Lung Squamous Cell Carcinoma Cells by Regulating miR-374a/b-5p/CADM2 Axis
Authors He J , Zhou X, Li L, Han Z
Received 26 September 2019
Accepted for publication 17 December 2019
Published 15 January 2020 Volume 2020:12 Pages 289—302
DOI https://doi.org/10.2147/CMAR.S232595
Checked for plagiarism Yes
Review by Single anonymous peer review
Peer reviewer comments 3
Editor who approved publication: Professor Seema Singh
Jia He, 1, 2 Xiaoyun Zhou, 1, 2 Li Li, 1, 2 Zhijun Han 1, 2
1Department of Thoracic Surgery, Peking Union Medical College Hospital, Beijing 100730, People’s Republic of China; 2Graduate School of Peking Union Medical College, Chinese Academy of Medical Sciences, Beijing 100730, People’s Republic of China
Correspondence: Zhijun Han
Department of Thoracic Surgery, Peking Union Medical College Hospital, Graduate School of Peking Union Medical College, Chinese Academy of Medical Sciences, No. 1 Shuaifuyuan, Dongcheng District, Beijing 100730, People’s Republic of China
Email [email protected]
Background: Lung squamous cell carcinoma (LUSC) accounts for approximately 30% of all lung cancers that possesses the highest occurrence and mortality in all cancer types. Long noncoding RNAs have been reported to modulate tumor development for several decades.
Aim of the Study: This research aims to investigate the role of MAGI2-AS3 in LUSC.
Methods: RT-qPCR tested genes (including MAGI2-AS3, miR-374a/b-5p and CADM2) expression. Cell proliferation was detected by colony formation and EdU assays. Cell migration and invasion were evaluated by transwell assay. Flow cytometry analysis of apoptotic cells and Western blot analysis on apoptosis-related genes were applied to measure cell apoptosis. Nuclear-cytoplasmic fractionation and FISH assay positioned MAGI2-AS3. The combination between miR-374a/b-5p and MAGI2-AS3 (or CADM2) was determined by luciferase reporter assay and RIP assay.
Results: MAGI2-AS3 inhibited the proliferative, migratory and invasive capability of LUSC cells with upregulated expression. Additionally, MAGI2-AS3 overexpression promoted cell apoptosis. We discovered that MAGI2-AS3 was located in the cytoplasm. Hereafter, we found out that MAGI2-AS3 targeted miR-374a/b-5p. CADM2 was targeted by miR-374a/b-5p. Finally, rescue assays indicated that the promoting effects of miR-374a/b-5p amplification on biological activities were restored by CADM2 addition.
Conclusion: In conclusion, lncRNA MAGI2-AS3 suppressed LUSC by regulating miR-374a/b-5p/CADM2 axis, which might potentially serve as a therapeutic marker for LUSC patients.
Keywords: lung squamous cell carcinoma, LUSC, MAGI2-AS3, miR-374a/b-5p, CADM2
Introduction
Lung cancer is one of the top 10 malignant tumors with increasing occurrence and mortality.1 Worse still, the incidence and mortality of lung cancer rank the first in all cancer types among the males and the second among the females.2 Small cell lung carcinoma and non-small-cell lung carcinoma (NSCLC) are the common subtypes of lung cancer. And NSCLC can be classified into lung adenocarcinoma (LUAD) and lung squamous cell carcinoma (LUSC).3,4 Known factors like smoking, air pollution and ionizing radiation are considered to be associated with the initiation and development of LUSC,5,6 but the pathology of LUSC remains unclear.
Long noncoding RNAs (lncRNAs) are a class of molecules with more than 200 nucleotides in length without capability encoding proteins.7 LncRNA dysregulation has been observed in various tumors.8,9 Specifically, downregulated lncRNAs repress tumor development and vice versa. As examples, HCG11 inhibits cell glioma growth by modulating miR-496/CPEB or miR-4425/MTA3 axis.10,11 Up-regulated HEIH promotes colorectal cancer tumorigenesis by cooperating with miR-939 to repress the transcription of Bcl-xl.12 Recently, MAGI2 antisense RNA 3 (MAGI2-AS3) is reported to act as a tumor suppressor in bladder cancer, breast cancer and hepatocellular carcinoma.13–15 Importantly, previous studies have identified that MAGI2-AS3 is down-regulated in NSCLC samples, including LUAD and LUSC samples.16,17 Moreover, we identified through GEPIA online tool based on TCGA data that MAGI2-AS3 was downregulated in LUSC samples versus normal samples. These findings indicated that MAGI2-AS3 might participate in LUSC. Also, Hao et al delineated that MAGI2-AS3 regulated NSCLC via miR-23a-3p/PTEN axis based on LUAD cell lines (A549, PC9, NCI-H441, and NCI-H1650).18 However, neither the biological function nor the regulatory mechanism of MAGI2-AS3 has been explored in LUSC before, which prompted us to investigate the role of MAGI2-AS3 in LUSC.
In mechanism, considerable evidence suggests that lncRNA is capable to regulate gene expression at the transcriptional level or post-transcriptional level.19,20 Additionally, the competitive endogenous RNA (ceRNA) pattern has attracted abundant attention. In this pattern, lncRNA enhances messenger RNA (mRNA) levels by sponging microRNA (miRNA).21,22 LINC00511 is reported to increase the E2F1 level by interacting with miR-185-3p in breast cancer.23 lncRNA XIST is supposed to modulate EZH2 expression via acting a molecular sponge of miR-101 in gastric cancer.24 Meanwhile, the regulatory mechanism of MAGI2-AS3 in LUSC remains uncharacterized.
To conclude, we attended to explore the biological function and regulatory mechanism of MAGI2-AS3 in LUSC and discovered that lncRNA MAGI2-AS3 suppressed several cellular processes of lung squamous cell carcinoma cells by regulating miR-374a/b-5p/CADM2 axis.
Materials and Methods
Tissue Samples
41 LUSC tissues and their paired adjacent noncancerous tissues were attained from patients in Peking Union Medical College Hospital by surgery excision between March 2013 and March 2014. No patients received radiotherapy or chemotherapy before surgery. Samples were frozen in liquid nitrogen at −80°C right after resection. Written informed consents were gained from all patients, with the approval of the Ethics Committee of Peking Union Medical College Hospital.
Cell Culture
Human bronchial epithelial cell (HBE) and LUSC cells (H2170, H226, SW900, SK-MES-1) were purchased from the American Type Culture Collection (ATCC; Manassas, VA, USA). In a humidified air with 5% CO2, cells were grown routinely at 37°C in RPMI-1640 medium (Gibco, Rockville, MD, USA) adding 1% penicillin/streptomycin (Invitrogen) and 10% fetal bovine serum (FBS; Gibco).
Cell Transfection
The pcDNA3.1/MAGI2-AS3, pcDNA3.1/CADM2 and the empty vectors were constructed by Genechem (Shanghai, China). The miR-374a-5p mimics, miR-374b-5p mimics, miR-23a-3p mimics and their corresponding NC mimics were constructed by GenePharma (Shanghai, China). Plasmids were individually transfected into SW900 or SK-MES-1 cells via Lipofectamine 3000 (Invitrogen, Carlsbad, CA, USA).
qRT-PCR
Through using TRIzol reagent (Invitrogen), total RNA was extracted. After reverse transcription into cDNA by using Evoscript Universal cDNA Master (Roche) (for lncRNA and mRNA) and Taqman MicroRNA reverse transcription kit (Applied Biosystems, Foster City, CA), qRT-PCR was implemented in ABI 7500 (Applied Biosystems, Foster City, CA, USA) adopting SYBR Green (Takara, Ohtsu, Japan) (for mRNA and lncRNA) and TaqMan MicroRNA Assays (Applied Biosystems) (for miRNA). Relative gene expression was normalized to U6 or GAPDH and fold changes were calculated through 2−ΔΔCt.
Colony Formation Assay
Transfected SW900 or SK-MES-1 cells were trypsinized and plated in 6-well culture plates (100 cells/well), followed by 14 days of incubation. Colonies were fixed and stained. Visible colonies consisting of more than 50 cells were counted, manually.
EdU Staining
Transfected SW900 or SK-MES-1 cells were inoculated into 96-well plates (8000 cells/well), followed by the addition of 50 μM EdU Reagent (RiboBio, Guangzhou, China) and incubation for 2 h. Cells were later subjected to Apollo (RiboBio) and DAPI (Sigma-Aldrich, St. Louis, MO, USA) in sequence. Images were taken by a fluorescence microscope (Nikon, Tokyo, Japan).
Flow Cytometry Analysis
Transfected SW900 or SK-MES-1 cells were washed twice using cold phosphate-buffered saline (PBS; Sigma-Aldrich) and later resuspended in 1 × binding buffer (Invitrogen). Cells were incubated for 15 min using Annexin V-FITC/PI (Invitrogen) away from light and at length analyzed through using the Accuri C6 flow cytometer (BD Biosciences, Mountain View, CA, USA).
Western Blot
Transfected SW900 or SK-MES-1 cells washed by cold PBS were lysed using RIPA buffer with an additional 1% protease inhibitor (Sigma-Aldrich) to extract protein. Evaluation of protein concentration was conducted by a BCA Protein Assay Kit (Thermo Fisher Scientific, Waltham, MA, USA). Protein was divided via SDS-PAGE gel (Bio-Rad, Hercules, CA, USA) and blotted onto PVDF membranes (Millipore, Billerica, MA, USA). Membranes blocked by 5% nonfat milk powder were incubated with primary antibodies against Cleaved caspase-3 (ab2302), total caspase-3 (ab13847), Bax (ab32503), Bcl-2 (ab32124), CADM2 (ab64873) and GAPDH (ab8245) which were bought from Abcam (Cambridge, MA, USA). Membranes were further incubated with secondary antibodies following washing by TBS-Tween 20 buffer (Sigma-Aldrich). Protein blots were subjected to ECL (Beyotime, Shanghai, China) and at last quantified with Image Lab software (Bio-Rad).
Transwell Assay
Migration and invasion capacities of transfected SW900 or SK-MES-1 cells were explored adopting transwell chambers (Corning, Cambridge, MA, USA). In migration assay, cells were added to the upper chambers in serum-free medium, whereas the lower chambers were added with medium supplying 10% FBS. After that, migratory cells were immobilized by 4% paraformaldehyde (PFA; Sigma-Aldrich), followed by being dyed in 0.1% crystal violet (Sigma-Aldrich). Migratory cells were counted via a microscope (Nikon). The upper chambers for invasion assay were pre-coated with Matrigel (BD Biosciences) and the other steps of invasion assay were similar to migration assay.
Subcellular Fractionation
Nuclear or cytoplasmic RNA in SW900 or SK-MES-1 cells was separated and purified with the application of a Cytoplasmic and Nuclear RNA Purification Kit (Norgen Biotek, Thorold, Canada). Expression patterns of MAGI2-AS3, GAPDH and U2 in nuclear and cytoplasm fractions were identified with qRT-PCR.
Fluorescence in situ Hybridization (FISH)
Fluorescently labeled MAGI2-AS3 probe was constructed by RiboBio. The Fluorescent in Situ Hybridization Kit (RiboBio) was employed for the detection of the signal of the probe. DAPI was utilized for counterstaining the nuclei of SW900 or SK-MES-1 cells. Images were acquired on a confocal microscope (Olympus, Tokyo, Japan).
RNA Immunoprecipitation (RIP)
The Magna-RIP RNA Binding Immunoprecipitation Kit (Millipore) was acquired for RIP. SW900 or SK-MES-1 cell lysates were acquired and incubated in the RIP reaction buffer. Anti-Ago2 antibody (Millipore) or anti-IgG antibody (Millipore) was conjugated to magnetic beads (Invitrogen). Enrichment of MAGI2-AS3, miR-374b-5p, miR-374a-5p or CADM2 was revealed using qRT-PCR.
Pulldown Assay
48 h after the LUSC cells were transfected with Bio-miR-374a-5p-WT, Bio-miR-374a-5p-Mut, Bio-miR-374b-5p-WT, Bio-miR-374b-5p-Mut, Bio-miR-23a-3p-WT, Bio-miR-23a-3p-Mut, or Bio-miR-NC, the cells were underwent PBS wash and were then incubated for 10 min in lysis buffer. RNA-protein complexes were excluded via blocking the magnetic beads in lysis buffer with the RNase-free BSA as well as yeast tRNA (Sigma). Then, incubate the lysates with streptavidin-coated magnetic beads (Life Technologies; Thermo Fisher Scientific, Waltham, MA, USA) for 4 h under 4°C, wash them twice using lysis buffer, three times with low-salt buffer, and then wash once using a high-salt buffer. Thereafter, bound RNAs were isolated applying the TRIzol reagent (Life Technologies). RNA expression was examined via qRT-PCR.
Luciferase Reporter Assay
The wild-type MAGI2-AS3 with the predictive interacting sequence of miR-374a/b-5p was cloned and constructed into the firefly luciferase psiCHECK-2 vector (Promega, Madison, WI, USA), termed as MAGI2-AS3-WT. Similarly, the mutant MAGI2-AS3 sequence was synthesized and inserted into the luciferase vector, termed as MAGI2-AS3-Mut. Besides, the CADM2 3ʹ-UTR plasmid containing wild-type or mutant miR-374a/b-5p or miR-23a-3p binding sites was inserted into the pmirGLO reporter vector (Promega). For luciferase analysis, both SW900 and SK-MES-1 cells were cultivated in 96-well plates and co-transfected with miR-374a/b-5p mimics or NC mimics using Lipofectamine 3000. After transfection for 48 h, cells were reaped and luciferase activity was determined in line with the instructions of dual-luciferase kit (Promega).
Animal Experiments
The BALB/c nude mice (male, 6-week-old) were bought from Beijing HFK Bioscience Co. Ltd. (Beijing, China). All mice were kept under pathogen-free conditions and the experiments followed Guidelines for Care and Use of Laboratory Animals and gained the approval of Peking Union Medical College Hospital. To monitor tumor growth, A549 cells (1×107) were transfected with pcDNA3.1 or pcDNA3.1/MAGI2-AS3 and then were subcutaneously injected into the mice. The tumor volume was monitored 4 days a time after the injection, and after 28 days, the mice were sacrificed. The volume was calculated following (length × width2)/2. The dissected tumors were weighed and subjected to immunohistochemistry using antibodies against Ki67 and PCNA (Abcam) following former description [20]. To monitor metastasis in vivo, transfected A549 cells were injected from tail vein of mice and the tumors dissected from mice were subjected to the staining of hematoxylin and eosin (H&E; Sigma-Aldrich).
Statistical Analysis
Experimental data from three independent assays were shown as mean ± SD through using Prism 6.0 (GraphPad, San Diego, CA, USA). Prognostic significance of MAGI2-AS3 was analyzed by Kaplan-Meier analysis and log-rank test. Gene expression correlations were assayed by Spearman correlation analysis. Differences were compared with Student’s t-test or one-way ANOVA. P < 0.05 indicated significant statistics.
Result
Knockdown of MAGI2-AS3 Inhibited the Proliferative, Migratory and Invasive Capability of LUSC Cells
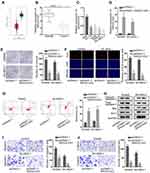
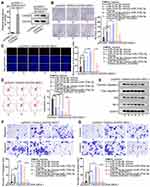
According to the data from the GEPIA database, MAGI2-AS3 was obviously down-regulated in tumor tissues in comparison with adjacent normal tissues (Figure 1A). Similarly, RT-qPCR assay also confirmed that LUSC tissues and cells presented lower MAGI2-AS3 levels than normal tissues and HBE cells, respectively (Figure 1B and C). Additionally, Kaplan-Meier analysis suggested that low MAGI2-AS3 expression was associated with low overall survival in LUSC patients (Figure S1A), indicating that MAGI2-AS3 bore prognostic significance in LUSC patients.
Since we have validated that SW900 and SK-MES-1 cells displayed the most significant down-regulation of the MAGI2-AS3 level among 4 LUSC cell lines, the two cell lines were chosen for in vitro gain-of-function assays. Subsequently, the MAGI2-AS3 level was prominently elevated by the transfection of pcDNA3.1/MAGI2-AS3 in SW900 and SK-MES-1 cells (Figure 1D). Then, we explored the specific function of MAGI2-AS3 in LUSC cells. Under the overexpression of MAGI2-AS3, colonies generated from SW900 cells were decreased from around 190 to around 70, and colonies generated by SK-MES-1 cells were decreased from around 160 to around 60 (Figure 1E). The number of EdU positive cells among SW900 and SK-MES-1 cells decreased from over 60 to about 20 due to MAGI2-AS3 overexpression (Figure 1F). According to flow cytometry analysis, the apoptosis ratio in LUSC cells increased from less than 5% to more than 10% due to MAGI2-AS3 overexpression (Figure 1G). Western blot assay disclosed that cleaved caspase-3 and Bax protein levels rose but Bcl-2 protein levels decreased in response to MAGI2-AS3 overexpression (Figure 1H). Additionally, transwell assay revealed that the migratory cells were decreased from nearly 150 to about 60 in SW900 cells and decreased from nearly 100 to about 20 in SK-MES-1 cells after MAGI2-AS3 overexpression (Figure 1I). The invaded cells decreased from over 100 to about 25 in SW900 cells and decreased from about 70 to about 20 in SK-MES-1 cells under MAGI2-AS3 overexpression (Figure 1J). To summarize, MAGI2-AS3 inhibited the proliferative, migratory and invasive capability of LUSC cells.
MAGI2-AS3 Sponged miR-374a/b-5p to Negatively Regulate Their Levels
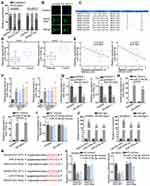
Mechanistically, the ceRNA pattern is widely reported as the way by which lncRNAs regulate gene expressions. Nuclear-cytoplasmic fractionation and FISH assay demonstrated that the majority of MAGI2-AS3 was distributed in the cytoplasm rather than the nucleus of LUSC cells (Figure 2A and B). Hence, we hypothesized that MAGI2-AS3 functioned as a ceRNA in LUSC. Bioinformatics analysis was adopted to seek potential miRNAs harboring binding site(s) in MAGI2-AS3. Based on Figure 2C, eight miRNAs were screened out (conditions: high stringency of CLIP Data and four cancer types of Pan-Cancer) after the prediction of starBase (http://starbase.sysu.edu.cn). Coincidently, miR-374a/b-5p was, respectively, reported to promote tumor development in breast cancer and hepatocellular carcinoma.15,25 Therefore, miR-374a-5p and miR-374b-5p were selected for further study. RT-qPCR suggested that miR-374a/b-5p levels in LUSC tissues were higher than that in normal tissues (Figure 2D). Spearman correlation analysis demonstrated that miR-374a/b-5p levels were in a negative correlation with MAGI2-AS3 expression (Figure 2E). High expression of miR-374a/b-5p was observed by RT-qPCR in LUSC cells, compared with normal cells (Figure 2F). Besides, forced MAGI2-AS3 expression decreased miR-374a/b-5p levels whereas MAGI2-AS3 displayed no differential expression in the context of miR-374a/b-5p overexpression (Figure 2G–I). According to RIP assay, MAGI2-AS3 and miR-374a/b-5p were co-immunoprecipitated by Ago2 antibody (the main component of RISC), implying that MAGI2-AS3 and miR-374a/b-5p were co-existed in RISC (Figure 2J). The binding sequences between MAGI2-AS3 and miR-374a/b-5p were predicted by starBase (Figure 2K). Luciferase reporter assays delineated that either miR-374a-5p or miR-374b-5p mimics triggered an obvious decline of luciferase activity of pmirGLO-MAGI2-AS3-WT whilst no significant changes were observed in MAGI2-AS3-Mut vector (Figure 2L). Taken together, MAGI2-AS3 targeted miR-374a/b-5p to negatively regulate their levels.
miR-374a/b-5p Directly Targeted CADM2 to Inhibit Its Activity
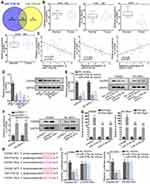
As shown in Figure 3A, the Venn diagram showed the possible target genes of miR-374a-5p (left) and miR-374b-5p (right) by PITA, miRmap, microT, miRanda and Pictar collected in Starbase website (http://starbase.sysu.edu.cn/) (Figure S1B). In detail, PARPB, LOLRAD4, CADM2 and RER1 were the common target genes of miR-374a-5p and miR-374b-5p. RT-qPCR indicated that only CADM2 was down-regulated in LUSC tissues (Figure 3B). Subsequently, the negative association between CADM2 and miR-374a-5p or miR-374b-5p, and the positive association between CADM2 and MAGI2-AS3 were displayed by Spearman correlation analysis (Figure 3C). LUSC cells also presented lower CADM2 mRNA and protein levels than HBE cells (Figure 3D). Moreover, both mRNA and protein levels of CADM2 were reduced by transfecting miR-374a-5p/miR-374b-5p mimics or pcDNA3.1/MAGI2-AS3 into SW900 and SK-MES-1 cells (Figure 3E and F). RIP assay manifested that CADM2 and miR-374a/b-5p were enriched in the anti-Ago2 group (Figure 3G). The binding sites of miR-374a-5p or miR-374b-5p on CADM2 were predicted and shown in Figure 3H. Based on luciferase reporter assay, the transfection of miR-374a-5p or miR-374b-5p mimics attenuated the luciferase activity of CADM2-WT vector but not that of CADM2-Mut (Figure 3I). In a word, MiR-374a/b-5p directly targeted CADM2.
MAGI2-AS3 Suppressed Several Cellular Processes of Lung Squamous Cell Carcinoma Cells by Regulating miR-374a/b-5p-CADM2 Axis
To prove whether MAGI2-AS3 acted as a tumor suppressor by modulating miR-374a/b-5p-CADM2 axis, we first detected the function of CADM2 on LUSC cells by overexpressing CADM2. CADM2 mRNA and protein expression were prominently upregulated by the transfection of pcDNA3.1/CADM2 (Figure 4A). We observed that overexpressing CADM2 reduced colonies generated by SW900 and SK-MES-1 cells (Figure S1C). The EdU positive cells decreased from more than 60 to less than 20 under CADM2 overexpression in 2 LUSC cell lines (Figure S1D). The apoptosis ratio was increased from less than 5% to more than 10% in 2 LUSC cell lines upon CADM2 overexpression (Figure S1E). The migrated cells and invaded cells were decreased under CADM2 ectopic expression in LUSC cells (Figure S1F and G). These data collectively indicated that CADM2 served as a tumor suppressor in LUSC cells.
Next, rescue assays were implemented to investigate whether miR-374a/b-5p were required for the regulation of MAGI2-AS3 on LUSC cells. According to the result of colony formation and EdU assays, cell proliferation presented an increase under miR-374a-5p (or miR-374b-5p) augmentation and such increase was further enhanced by the transfection of miR-374a-5p mimics and miR-374b-5p mimics, and then abrogated by CADM2 overexpression (Figure 4B and C). According to flow cytometry, the apoptotic ratio in pcDNA3.1/MAGI2-AS3-transfected SK-MES-1 cells reduced to around 7% by respective overexpression of miR-374a-5p and miR-374b-5p, and the apoptotic ratio further dropped to less than 5% as a result of miR-374a/b-5p co-overexpression. Later, the apoptotic ratio was recovered to over 10% by CADM2 overexpression (Figure 4D). Over and above that, the transfection of pcDNA3.1/CADM2 offset the effect of miR-374a/b-5p overexpression on reducing cleaved caspase-3 or Bax protein expressions and inducing Bcl-2 protein expression (Figure 4E). Correspondingly, the promoting effect of miR-374a-5p and miR-374b-5p overexpression on cell migration and invasion was rescued by the transfection of pcDNA3.1/CADM2 (Figure 4F and G). All in all, MAGI2-AS3 suppressed the proliferation and migration of LUSC cells via miR-374a/b-5p/CADM2 axis.
MAGI2-AS3/miR-23b-3p/PTEN Axis Functioned in LUSC Cells
According to former studies, MAGI2-AS3 also regulated other target genes via miR-374a/b-5p to exhibit is tumor-suppressing effect. Notably, MAGI2-AS3 regulated miR-374b-5p/SMG1 axis in hepatocellular carcinoma,15 MAGI2-AS3 regulated miR-374a/PTEN axis in breast cancer.26 Hence, we tried to detect whether SMG1 and PTEN were alternate targets for MAGI2-AS3/miR-374a/b-5p axis. Interestingly, RT-qPCR data depicted that SMG1 presented no significant change in LUSC samples versus the adjacent normal samples (Figure S2A). However, PTEN was significantly downregulated in LUSC samples (Figure S2B), indicating that PTEN, rather than SMG1, was involved in LUSC. Unexpectedly, pulldown assay showed that neither miR-374a-5p nor miR-374b-5p interacted with PTEN mRNA in LUSC cells (Figure S2C). Also, miR-374a-5p mimic or miR-374b-5p mimic failed to inhibit PTEN mRNA and protein expressions in LUSC cells (Figure S2D). Instead, we discovered that MAGI2-AS3 overexpression induced PTEN mRNA and protein expressions in LUSC cells (Figure S2E), indicating that MAGI2-AS3 might regulate PTEN expression independent from miR-374a/b-5p. A previous report validated that MAGI2-AS3 can regulate PTEN expression via miR-23a-3p in NSCLC by using LUAD cell lines.18 Therefore, we wondered whether MAGI2-AS3 could regulate PTEN via miR-23a-3p in LUSC cells as well. Expectedly, pulldown assay confirmed the enrichment of PTEN mRNA and MAGI2-AS3 in the bio-miR-23a-2p-WT group rather than bio-miR-NC and bio-miR-23a-3p-Mut group (Figure S2F). RIP assay showed the co-immunoprecipitation of MAGI2-AS3, miR-23a-3p, and PTEN mRNA in the Ago2 group (Figure S2G). We also obtained the binding sites on PTEN for miR-23a-3p via TargetScan (http://www.targetscan.org/vert_72/) (Figure S2H). Luciferase activity of PTEN (WT), rather than PTEN (Mut) was decreased by miR-23a-3p mimic in 2 LUSC cell lines (Figure S2H). Together, it was suggested that MAGI2-AS3 regulated PTEN via miR-23a-3p in LUSC cells. Moreover, we verified that miR-23a-3p was upregulated in LUSC samples versus the adjacent normal samples (Figure S3A). We also demonstrated the negative correlation of miR-23a-3p with MAGI2-AS3 and PTEN and the positive correlation between MAGI2-AS3 and PTEN in LUSC samples (Figure S3B).
Thereafter, rescue assays were conducted to validate the function of MAGI2-AS3/miR-23b-3p/PTEN axis in LUSC cells. We confirmed via Western blot analysis that overexpressing MAGI2-AS3 induced PTEN level, whereas the induction of PTEN was reversed by miR-23a-3p mimic, and overexpressing PTEN restored the PTEN level reduced by miR-23a-3p mimic (Figure S3C). miR-23a-3p mimic rescued the colony formation and EdU positive LUSC cells reduced by MAGI2-AS3 overexpression, and the effect of miR-23a-3p mimic was reversed by PTEN overexpression in LUSC cells (Figure S3D and E). Overexpression of miR-23a-3p mimic counteracted the high apoptosis ratio in LUSC cells under MAGI2-AS3 overexpression, and PTEN overexpression reversed the effect of miR-23a-3p mimic (Figure S3F). The migration and invasion of LUSC cells were decreased by MAGI2-AS3 overexpression and restored by miR-23a-3p mimic, and the restoration of migration and invasion was later counteracted by PTEN overexpression (Figure S3G and H). Jointly, MAGI2-AS3 also suppressed proliferation, migration, and invasion via miR-23a-3p/PTEN axis in LUSC cells.
Function of MAGI2-AS3 on LUSC Tumorigenesis and Metastasis in vivo
Finally, we carried out in vivo assays to investigate the function of MAGI2-AS3 in LUSC. SK-MES-1 cells transfected with pcDNA3.1 or pcDNA3.1/MAGI2-AS3 were injected into nude mice and the tumor growth was observed. As a result, MAGI2-AS3 overexpression retarded the tumor growth and reduced tumor weight in mice (Figure 5A and B). The levels of MAGI2-AS3, CAMD2, and PTEN were increased in xenografts with MAGI2-AS3 overexpression (Figure 5C). Western blot analysis showed that MAGI2-AS3 overexpression induced the levels of CADM2, PTEN, c-Caspase 3, and Bax, and reduced the level of Bcl-2 (Figure 5D). The IHC staining ratio of proliferation index (Ki67 and PCNA) in xenografts of mice decreased upon the overexpression of MAGI2-AS3 (Figure 5E). Moreover, HE staining depicted that the metastatic nodules in mice decreased under MAGI2-AS3 overexpression (Figure 5F). Together, these results indicated that MAGI2-AS3 retarded tumorigenesis and metastasis in vivo.
Discussion
LUSC, accounting for 30% of all lung cancer types, is a horrible threat to individual life.27,28 Modern advanced methods like surgery, chemotherapy and radiotherapy have been applied to treat LUSC.28,29 Nevertheless, LUSC patients still present a dismal prognosis with a low overall 5-year survival rate due to tumor metastasis and recurrence.30,31 Hence, further exploration of carcinogenesis is essential to the identification of potentially diagnostic or therapeutic targets.
LncRNAs can be classified into genic (exonic, intronic, overlapping) and intergenic lncRNAs according to their location with respect to the nearest protein-coding transcripts.19 Functionally, a multitude of researches claim that lncRNA may serve as important regulatory factors in diverse biological processes including cell proliferation, differentiation, migration, invasion or apoptosis.32,33 Long noncoding RNA ZFAS1 is proved to improve cell proliferation and invasion by sponging miR-484 in colorectal cancer.34 The long noncoding RNA SNHG14 represses cell proliferation but boosts cell apoptosis of glioma cells by sponging miR-92a-3p.35 MAGI2-AS3 has been reported as an anti-tumor lncRNA in several cancers, including hepatocellular cancer and breast cancer.15,26 The downregulation of MAGI2-AS3 has been proved in NSCLC including LUAD and LUSC.16,17 A recent research validated the tumor-suppressive role of MAGI2-AS3 in NSCLC based on LUAD cell lines.18 However, the role of MAGI2-AS3 in LUSC is yet to be shown. We identified the downregulation of MAGI2-AS3 in LUSC samples based on TCGA data from GEPIA and validated such result in LUSC samples by RT-qPCR, suggesting the participation of MAGI2-AS3 in LUSC. Importantly, we were the first to investigate the function of MAGI2-AS3 in LUSC in vitro and in vitro. We demonstrated the MAGI2-AS3 inhibited proliferation, induced apoptosis, and increased migration and invasion in vitro, and proved that MAGI2-AS3 overexpression attenuated tumorigenesis and metastasis in vivo.
Mechanistically, lncRNA interacts with miRNA to modulate the target gene(s) level. Particularly, lncRNA generally acts as a ceRNA in the cytoplasm of cells. Based on this hypothesis, we conjectured that MAGI2-AS3 might also function in this pattern. Coincidently, we found that the majority of MAGI2-AS3 was distributed in the cytoplasm of LUSC cells. Additionally, MAGI2-AS3 negatively regulated miR-374a/b-5p levels while MAGI2-AS3 displayed no changes in the context of miR-374a/b-5p overexpression. Then, the combination between MAGI2-AS3 and miR-374a/b-5p was confirmed. Hereafter, cell adhesion molecule 2 (CADM2) was chosen as the target gene of miR-374a/b-5p, which has been reported to be down-regulated in glioma and hepatocellular carcinoma.36,37 We first validated that miR-374a/b-5p negatively modulated mRNA and protein levels of CADM2 by binding to CAMD2 mRNA. Rescue assays revealed that the promoting effects resulted from miR-374a/b-5p amplification on the proliferation, apoptosis resistance, migration and invasion were retarded by transfection of pcDNA3.1/CADM2.
Moreover, several former works have shown that MAGI2-AS3 can target other genes via miR-374a-5p and miR-374b-5p, such as PTEN and SMG1 in breast and hepatocellular cancers.15,26 Interestingly, we discovered that SMG1 presented no significant alteration in LUSC samples whereas PTEN was downregulated in LUSC samples, indicating the involvement of PTEN in LUSC. However, we showed that unlike in breast cancer and hepatocellular cancer cells, miR-374a/b-5p cannot interact with and affect PTEN in LUSC cells. Instead, MAGI2-AS3 can still upregulate PTEN in LUSC cells. According to a former study, MAGI2-AS3 targeted miR-23a-3p in LUAD cells to upregulate PTEN.18 In concordance, we firstly validated in LUSC cells that MAGI2-AS3 also regulated miR-23a-3p/PTEN axis to suppress cell proliferation, migration, and invasion. Therefore, it was suggested that.
All in all, for the first time, we verified the function and mechanism of MAGI2-AS3 in LUSC by demonstrating a novel ceRNA network MAGI2-AS3/miR-374a/b-5p/CAMD1 in LUSC and validating that anti-cancer role of MAGI2-AS3/miR-23a-3p/PTEN axis reported in LUAD was also effective in LUSC. This may provide a novel insight into the treatment of LUSC patients. Yet, it was just the initial exploration of MAGI2-AS3 and other related mechanisms in LUSC remain to be explored in the future.
Ethics Approval and Informed Consent
This study was carried out in accordance with the principles of the Declaration of Helsinki and was approved by the Ethics Committee of Peking Union Medical College Hospital. The written informed consents were gained from all patients.
Consent for Publication
All authors agreed to publish it.
Data Sharing Statement
Research data are not shared.
Acknowledgment
Thanks a lot to all individuals or teams that were involved in our research.
Author Contributions
Jia He: Conceptualization, Data curation, Formal analysis, Project administration. Xiaoyun Zhou: Investigation, Methodology, Resources. Li Li: Software, Supervision, Validation, Visualization. Zhijun Han: Writing – original draft, Writing – review and editing. All authors contributed to data analysis, drafting or revising the article, gave final approval of the version to be published, and agree to be accountable for all aspects of the work.
Disclosure
The authors declare that there are no competing interests in this study.
References
1. Mao Y, Yang D, He J, Krasna MJ. Epidemiology of lung cancer. Surg Oncol Clin N Am. 2016;25(3):439–445. doi:10.1016/j.soc.2016.02.001
2. Evans M. Lung cancer: needs assessment, treatment and therapies. Br J Nurs. 2013;22(17):
3. Collins LG, Haines C, Perkel R, Enck RE. Lung cancer: diagnosis and management. Am Fam Physician. 2007;75(1):56–63.
4. Qiu ZW, Bi JH, Gazdar AF, Song K. Genome-wide copy number variation pattern analysis and a classification signature for non-small cell lung cancer. Genes Chromosomes Cancer. 2017;56(7):559–569. doi:10.1002/gcc.22460
5. Rivera GA, Wakelee H. Lung cancer in never smokers. Adv Exp Med Biol. 2016;893:43–57.
6. Osmani L, Askin F, Gabrielson E, Li QK. Current WHO guidelines and the critical role of immunohistochemical markers in the subclassification of non-small cell lung carcinoma (NSCLC): moving from targeted therapy to immunotherapy. Semin Cancer Biol. 2018;52(Pt 1):103–109. doi:10.1016/j.semcancer.2017.11.019
7. Xing Z, Park PK, Lin C, Yang L. LncRNA BCAR4 wires up signaling transduction in breast cancer. RNA Biol. 2015;12(7):681–689. doi:10.1080/15476286.2015.1053687
8. Thin KZ, Liu X, Feng X, Raveendran S, Tu JC. LncRNA-DANCR: a valuable cancer related long non-coding RNA for human cancers. Pathol Res Pract. 2018;214(6):801–805. doi:10.1016/j.prp.2018.04.003
9. Luo D, Deng B, Weng M, Luo Z, Nie X. A prognostic 4-lncRNA expression signature for lung squamous cell carcinoma. Artif Cells Nanomed Biotechnol. 2018;46(6):1207–1214. doi:10.1080/21691401.2017.1366334
10. Chen Y, Bao C, Zhang X, Lin X, Huang H, Wang Z. Long non-coding RNA HCG11 modulates glioma progression through cooperating with miR-496/CPEB3 axis. Cell Prolif. 2019;52(5):e12615.
11. Zhang L, Cao Y, Kou X, et al. Long non-coding RNA HCG11 suppresses the growth of glioma by cooperating with the miR-4425/MTA3 axis. J Gene Med. 2019;21(4):e3074. doi:10.1002/jgm.v21.4
12. Cui C, Zhai D, Cai L, Duan Q, Xie L, Yu J. Long noncoding RNA HEIH promotes colorectal cancer tumorigenesis via counteracting miR-939Mediated transcriptional repression of Bcl-xL. Cancer Res Treat. 2018;50(3):992–1008. doi:10.4143/crt.2017.226
13. Wang F, Zu Y, Zhu S, et al. Long noncoding RNA MAGI2-AS3 regulates CCDC19 expression by sponging miR-15b-5p and suppresses bladder cancer progression. Biochem Biophys Res Commun. 2018;507(1–4):231–235. doi:10.1016/j.bbrc.2018.11.013
14. Yang Y, Yang H, Xu M, et al. Long non-coding RNA (lncRNA) MAGI2-AS3 inhibits breast cancer cell growth by targeting the Fas/FasL signalling pathway. Hum Cell. 2018;31(3):232–241. doi:10.1007/s13577-018-0206-1
15. Yin Z, Ma T, Yan J, et al. LncRNA MAGI2-AS3 inhibits hepatocellular carcinoma cell proliferation and migration by targeting the miR-374b-5p/SMG1 signaling pathway. J Cell Physiol. 2019;234(10):18825–18836. doi:10.1002/jcp.28521
16. Luo CL, Xu ZG, Chen H, et al. LncRNAs and EGFRvIII sequestered in TEPs enable blood-based NSCLC diagnosis. Cancer Manag Res. 2018;10:1449–1459. doi:10.2147/CMAR.S164227
17. Huang J, Li Y, Lu Z, et al. Analysis of functional hub genes identifies CDC45 as an oncogene in non-small cell lung cancer - a short report. Cell Oncol (Dordr). 2019;42(4):571–578. doi:10.1007/s13402-019-00438-y
18. Hao XZ, Yang K. LncRNA MAGI2-AS3 suppresses the proliferation and invasion of non-small cell lung carcinoma through miRNA-23a-3p/PTEN axis. Eur Rev Med Pharmacol Sci. 2019;23(17):7399–7407. doi:10.26355/eurrev_201909_18848
19. Xu J, Bai J, Zhang X, et al. A comprehensive overview of lncRNA annotation resources. Brief Bioinform. 2017;18(2):236–249. doi:10.1093/bib/bbw015
20. Jathar S, Kumar V, Srivastava J, Tripathi V. Technological developments in lncRNA biology. Adv Exp Med Biol. 2017;1008:283–323.
21. Li LJ, Zhao W, Tao SS, et al. Competitive endogenous RNA network: potential implication for systemic lupus erythematosus. Expert Opin Ther Targets. 2017;21(6):639–648. doi:10.1080/14728222.2017.1319938
22. Zheng L, Xiang C, Li X, et al. STARD13-correlated ceRNA network-directed inhibition on YAP/TAZ activity suppresses stemness of breast cancer via co-regulating hippo and Rho-GTPase/F-actin signaling. J Hematol Oncol. 2018;11(1):72. doi:10.1186/s13045-018-0613-5
23. Küffer S, Gutting T, Belharazem D, et al. Insulin‐like growth factor 2 expression in prostate cancer is regulated by promoter‐specific methylation. Mol Oncol. 2018;12(2):256–266. doi:10.1002/mol2.2018.12.issue-2
24. Chen DL, Ju HQ, Lu YX, et al. Long non-coding RNA XIST regulates gastric cancer progression by acting as a molecular sponge of miR-101 to modulate EZH2 expression. J Exp Clin Cancer Res. 2016;35(1):142. doi:10.1186/s13046-016-0420-1
25. Son D, Kim Y, Lim S, et al. miR-374a-5p promotes tumor progression by targeting ARRB1 in triple negative breast cancer. Cancer Lett. 2019;454:224–233. doi:10.1016/j.canlet.2019.04.006
26. Du S, Hu W, Zhao Y, et al. Long non-coding RNA MAGI2-AS3 inhibits breast cancer cell migration and invasion via sponging microRNA-374a. Cancer Biomark. 2019. doi:10.3233/CBM-182216
27. Chen M, Liu X, Du J, Wang XJ, Xia L. Differentiated regulation of immune-response related genes between LUAD and LUSC subtypes of lung cancers. Oncotarget. 2017;8(1):133–144. doi:10.18632/oncotarget.13346
28. Yang C, Zheng J, Xue Y, et al. The effect of MCM3AP-AS1/miR-211/KLF5/AGGF1 axis regulating glioblastoma angiogenesis. Front Mol Neurosci. 2017;10:437. doi:10.3389/fnmol.2017.00437
29. Ma Z, Wang Y, He B, et al. Expression of miR-590 in lung cancer and its correlation with prognosis. Oncol Lett. 2018;15(2):1753–1757. doi:10.3892/ol.2017.7497
30. Wei Y, Zhang X. Transcriptome analysis of distinct long non-coding RNA transcriptional fingerprints in lung adenocarcinoma and squamous cell carcinoma. Tumour Biol. 2016. doi:10.1007/s13277-016-5422-2
31. Miyoshi A, Kanao S, Naoi H, Otsuka H, Yokoi T. Investigation of the clinical features of lower uterine segment carcinoma: association with advanced stage disease and indication of poorer prognosis. Arch Gynecol Obstet. 2018;297(1):193–198. doi:10.1007/s00404-017-4576-5
32. Wang MW, Liu J, Liu Q, et al. LncRNA SNHG7 promotes the proliferation and inhibits apoptosis of gastric cancer cells by repressing the P15 and P16 expression. Eur Rev Med Pharmacol Sci. 2017;21(20):4613–4622.
33. Long B, Li N, Xu XX, et al. Long noncoding RNA FTX regulates cardiomyocyte apoptosis by targeting miR-29b-1-5p and Bcl2l2. Biochem Biophys Res Commun. 2018;495(1):312–318. doi:10.1016/j.bbrc.2017.11.030
34. Xie S, Ge Q, Wang X, Sun X, Kang Y. Long non-coding RNA ZFAS1 sponges miR-484 to promote cell proliferation and invasion in colorectal cancer. Cell Cycle. 2018;17(2):154–161. doi:10.1080/15384101.2017.1407895
35. Wang Q, Teng Y, Wang R, et al. The long non-coding RNA SNHG14 inhibits cell proliferation and invasion and promotes apoptosis by sponging miR-92a-3p in glioma. Oncotarget. 2018;9(15):12112–12124. doi:10.18632/oncotarget.23960
36. Li D, Zhang Y, Zhang H, et al. CADM2, as a new target of miR-10b, promotes tumor metastasis through FAK/AKT pathway in hepatocellular carcinoma. J Exp Clin Cancer Res. 2018;37(1):46. doi:10.1186/s13046-018-0699-1
37. Liu N, Yang C, Bai W, et al. CADM2 inhibits human glioma proliferation, migration and invasion. Oncol Rep. 2019;41(4):2273–2280. doi:10.3892/or.2019.7010
 © 2020 The Author(s). This work is published and licensed by Dove Medical Press Limited. The full terms of this license are available at https://www.dovepress.com/terms.php and incorporate the Creative Commons Attribution - Non Commercial (unported, v3.0) License.
By accessing the work you hereby accept the Terms. Non-commercial uses of the work are permitted without any further permission from Dove Medical Press Limited, provided the work is properly attributed. For permission for commercial use of this work, please see paragraphs 4.2 and 5 of our Terms.
© 2020 The Author(s). This work is published and licensed by Dove Medical Press Limited. The full terms of this license are available at https://www.dovepress.com/terms.php and incorporate the Creative Commons Attribution - Non Commercial (unported, v3.0) License.
By accessing the work you hereby accept the Terms. Non-commercial uses of the work are permitted without any further permission from Dove Medical Press Limited, provided the work is properly attributed. For permission for commercial use of this work, please see paragraphs 4.2 and 5 of our Terms.