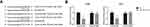Back to Journals » Cancer Management and Research » Volume 12
Long Non-Coding RNA TTN-AS1 Serves as a Competing Endogenous RNA of miR-195 to Facilitate Clear Cell Renal Cell Carcinoma Progression
Authors Lin K, Chen H, Su C, Zhu H, Lai C, Shi Y
Received 12 February 2020
Accepted for publication 1 April 2020
Published 4 May 2020 Volume 2020:12 Pages 3091—3097
DOI https://doi.org/10.2147/CMAR.S249456
Checked for plagiarism Yes
Review by Single anonymous peer review
Peer reviewer comments 2
Editor who approved publication: Professor Yong Teng
Keng Lin,1 Hao Chen,2 Chunyan Su,3 Huanjin Zhu,1 Changchun Lai,4 Yaling Shi5
1Clinical Laboratory, Maternal and Children Health Care Hospital (Huzhong Hospital) of Huadu, Guangzhou 510800, People’s Republic of China; 2Clinical Laboratory, The Affiliated Oncology Hospital of Sun Yat-sen University, Guangzhou 510000, People’s Republic of China; 3Clinical Laboratory, The Second People’s Hospital of Gaozhou, Gaozhou 525200, People’s Republic of China; 4Clinical Laboratory, People’s Hospital of Maoming, Maoming 525000, People’s Republic of China; 5Clinical Laboratory, Guangzhou Eighth People’s Hospital, Guangzhou Medical University, Guangzhou 510000, People’s Republic of China
Correspondence: Yaling Shi
Clinical Laboratory, Guangzhou Eighth People’s Hospital, Guangzhou Medical University, Guangzhou 510060, People’s Republic of China
Email [email protected]
Introduction: Clear cell renal cell carcinoma (ccRCC) is an aggressive human malignancy. Long non-coding RNAs (lncRNAs) are critical regulators in many malignant tumors, including ccRCC. The aim of this study is to investigate the expression, functions and molecular mechanisms of lncRNA TTN-AS1 in ccRCC.
Methods: A total of 145 paired cancer and normal tissues were collected from patients with ccRCC. The expression levels of TTN-AS1 and miR-195 in the tissues or cells were measured by RT-qPCR analysis. The expression levels of cyclin D1 protein were measured by Western blot analysis. Cell proliferation and cell cycle distribution were detected by MTT assay and flow cytometer analysis, respectively. The binding relationship between miR-195 and TTN-AS1 or cyclin D1 mRNA was validated by dual-luciferase reporter assay.
Results: Our results revealed that TTN-AS1 expression levels in human ccRCC tissues and cell lines were markedly increased. High expression of TTN-AS1 was closely associated with adverse clinicopathological characteristics of ccRCC patients. Gain- and loss-of-function experiments showed that TTN-AS1 overexpression promoted the proliferation and cell cycle transition of ccRCC cells, while the malignant traits were obviously suppressed after TTN-AS1 knockdown. Mechanistically, miR-195 was found to bind with and to be negatively regulated by TTN-AS1 in ccRCC cells. Further, we showed that cyclin D1 is a direct target of miR-195 in ccRCC, and rescue assays verified that restoration of miR-195 expression partially blocked the oncogenic effects of TTN-AS1 in ccRCC cells.
Conclusion: Our study provides a novel mechanism of TTN-AS1/miR-195/cyclin D1 regulatory axis in ccRCC, which may become a breakthrough for ccRCC therapy in the future.
Keywords: clear cell renal cell carcinoma, long non-coding RNA TTN-AS1, miR-195, cell cycle, cyclin D1
Introduction
Renal cell carcinoma (RCC), originating in the renal cortex, is one of the most common malignancies of genitourinary system, and clear cell RCC (ccRCC) accounts for approximately 80% of all RCC cases.1 Approximately 1/3 of ccRCC patients have localized and distant metastasis or recurrence after surgical resection, and the long-term prognosis for these patients remains dismal.2 Accordingly, it is extremely necessary to better understand the molecular mechanisms of ccRCC, and identify potential therapeutic targets for this aggressive malignancy.
Long non-coding RNAs (lncRNAs) are a critical group of non-protein coding RNA transcripts that have a length greater than 200 nucleotides.3 LncRNAs were previously considered as “transcriptional junk”, but at present, increasing number of studies have illustrated their important regulatory roles in diverse human diseases, ranging from neurodegeneration to cancer.4,5 Among a wide variety of cancer-related lncRNAs, TTN antisense RNA 1 (TTN-AS1) was recently shown to be overexpressed in many malignant tumors, such as gastric cancer, lung adenocarcinoma and colorectal cancer.6–9 However, the clinical significance of TTN-AS1 in ccRCC and the related regulatory functions remain elusive.
Therefore, in this study, we carried out a series of experiments to investigate the expression, functions and molecular mechanisms of TTN-AS1 in ccRCC. Our results may contribute to a better understanding of ccRCC progression.
Materials and Methods
Tissue Sample Collection
ccRCC tissues and their adjacent normal tissues were collected from 145 patients who underwent primary surgical resection from April 2017 to January 2020 at The Affiliated Oncology Hospital of Sun Yat-sen University (Guangzhou, China) and Guangzhou Eighth People’s Hospital (Guangzhou, China). None of patients had received radiotherapy or chemotherapy before surgery. All samples were immediately snap-frozen in liquid nitrogen and stored at −80 °C until further use. Our study was approved by the Ethics Committee of The Affiliated Oncology Hospital of Sun Yat-sen University and the Ethics Committee of Guangzhou Eighth People’s Hospital. Written informed consent was obtained from all participants.
Cell Lines
Three human ccRCC cell lines, including ACHN, 786-O, SN12-PM6, and one normal human kidney proximal tubular epithelial cell line HK-2, obtained from Shanghai Cell Bank (Shanghai, China), were cultured in Dulbecco’s modified Eagle’s medium (DMEM; Hyclone, Logan, UT, USA) supplemented with 10% fetal bovine serum (FBS; Hyclone), penicillin (100 U/mL) and streptomycin (100 mg/mL) at 37°C in a humidified incubator with 5% CO2.
Cell Transfection
The small interfering RNA (siRNA) targeting TTN-AS1 (si-TTN-AS1), siRNA negative control (si-NC), miR-195 mimics, and scrambled oligonucleotides (NC) were all obtained from Shanghai GenePharma Co., Ltd. (Shanghai, China). The cDNA encoding TTN-AS1 was amplified by PCR, and then ligated into pcDNA3.1 ( + ) vector (Invitrogen). Cell transfection was performed using Lipofectamine 2000 (Invitrogen). After 48 h, the transfection efficiency was validated by RT-qPCR analysis.
RNA Extraction and RT-qPCR Analysis
Total RNA was isolated using TRIzol Reagent (Invitrogen). Cytoplasm and nuclear RNA of cells were separated and extracted using the PARIS Kit (Invitrogen). First strand cDNA was then generated using the PrimerScript™ reagent Kit (TaKaRa, Dalian, China). PCR amplification was carried out using a SYBR Green PCR kit (TaKaRa) on an ABI PRISM 7300 Sequence Detection system (Applied Biosystems, Foster City, CA, USA). Relative quantification of gene expression was calculated by the 2−ΔΔCt method,10 with GAPDH or U6 as an endogenous control. All experiments were repeated in independent triplicate.
MTT Assay
Cells were seeded into 96-well plates at a density of 4×103/well. At the end of each period, 20 μL of MTT (5 mg/mL; Sigma-Aldrich, St. Louis, MO, USA) was added into each well. After incubation at 37°C for another 4 h, the culture medium was removed, and 150 μL of DMSO (Sigma-Aldrich) was added. Absorbance was measured at 570 nm by a microplate reader (MultiskanEX, Lab systems, Helsinki, Finland). All experiments were repeated in independent triplicate.
Cell Cycle Analysis
Cells were fixed with ice-cold 70% ethanol overnight and then stained with 20 μg/mL propidium iodide using the Cycletest™ Plus DNA Reagent Kit (BD Biosciences, San Jose, CA, USA). After incubation for 1 h at 37°C in the dark, the stained cells were collected and then subjected to a flow cytometer (FACSCalibur; BD Biosciences). All experiments were repeated in independent triplicate.
Western Blot Analysis
Protein lysates were extracted using radio immunoprecipitation assay (RIPA) lysis buffer (Beyotime, Shanghai, China), fractionated by SDS-PAGE gel and then electrophoretically transferred to polyvinylidene difluoride (PVDF) membranes (Millipore, Bedford, MA, USA). The membranes were blocked in 5% non-fat dried milk for 2 h at room temperature and then incubated with primary antibodies at 4 °C overnight. Afterwards, the membranes were incubated with HRP-conjugated secondary antibody for 1 h at room temperature. The blot signals were visualized with an enhanced chemiluminescence kit (GE Healthcare, Little Chalfont, UK). GAPDH was used as an internal control. All experiments were repeated in independent triplicate.
Dual-Luciferase Reporter Assay
The fragment of TTN-AS1 or cyclin D1 mRNA containing miR-195 binding sites was amplified by PCR and inserted into the psiCHECK-2 luciferase reporter vector (Promega, Madison, WI, USA). The binding sites were mutated by the Quick-change site-directed mutagenesis kit (Agilent Technologies, Santa Clara, CA, USA). The reporter plasmid was transfected into cells in the presence of miR-195 mimics or NC using Lipofectamine 2000. At 48 h after transfection, the luciferase activities were measured using the Dual-Luciferase Reporter Assay System (Promega). All experiments were repeated in independent triplicate.
Statistical Analysis
Experimental data were expressed as mean ± standard deviation (SD). All statistical analyses were conducted by using GraphPad Prism version 6.0 (GraphPad Software, Inc., San Diego, CA, USA) and SPSS 18.0 software (SPSS Inc., Chicago, IL, USA). Group difference was assessed using Student’s t-test or one-way ANOVA. A P value < 0.05 was considered as statistically significant.
Results
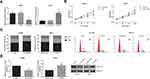
TTN-AS1 Is Upregulated in ccRCC
Through RT-qPCR analysis, we first observed that TTN-AS1 expression was notably elevated in ccRCC tissues than that in adjacent normal tissues (Figure 1A). We then divided all the patients into two groups according to the mean TTN-AS1 expression: high expression group (n=65) and low expression group (n=80), and we observed that high level of TTN-AS1 expression in ccRCC patients was significantly correlated with larger tumor size (P=0.021) and advanced TNM stage (P=0.008) (Table 1). In addition, as shown in Figure 1B, in all tested ccRCC cell lines (ACHN, 786-O, SN12-PM6), TTN-AS1 expression was significantly higher than that of normal HK-2 cell line. ACHN cells (highest TTN-AS1 expression) and 786-O cells (lowest TTN-AS1 expression) were chosen for further experiments.
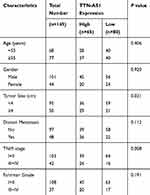 |
Table 1 Correlation Between TTN-AS1 Expression and Clinicopathological Characteristics in ccRCC Patients |
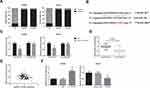
TTN-AS1 Promotes ccRCC Cell Proliferation and Cell Cycle Progression
To manipulate the expression of TTN-AS1 in ccRCC cells, si-TTN-AS1 was introduced into ACHN cells, and we noticed a significant reduction of TTN-AS1 expression (Figure 2A). Besides, TTN-AS1 was overexpressed in 786-O cells by transfection with pcDNA3.1-TTN-AS1. We then performed MTT assay to explore the effects of TTN-AS1 on ccRCC cell proliferation. As exhibited in Figure 2B, the proliferation rate of ACHN cells was tremendously impaired when TTN-AS1 was knocked down, whereas overexpression of TTN-AS1 could otherwise enhance the proliferation in 786-O cells. Besides, cell cycle analysis confirmed that cell cycle progression was markedly suppressed by TTN-AS1 knockdown in ACHN cells, while 786-O cells with overexpressing TTN-AS1 had a decreased G0/G1 population (Figure 2C). Western blot analysis further showed that the expression level of cyclin D1 protein was decreased by TTN-AS1 knockdown in ACHN cells, as well as increased by TTN-AS1 overexpression in786-O cells (Figure 2D).
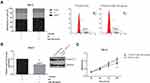
TTN-AS1 Works as a ceRNA of miR-195 in ccRCC
As revealed by Figure 3A, TTN-AS1 was distributed mostly in the cytoplasm of ACHN and 786-O cells, and through the Starbase database (http://starbase.sysu.edu.cn/index.php), we identified the potential binding sites for miR-195 on TTN-AS1 (Figure 3B). Then, dual-luciferase reporter assay showed that the luciferase activity of TTN-AS1-WT but not the TTN-AS1-MUT was reduced strikingly by co-transfection with miR-195 mimics in both ACHN and 786-O cells (Figure 3C). Furthermore, we showed that miR-195 expression was significantly decreased (Figure 3D) and inversely correlated with TTN-AS1 expression in the ccRCC tissues (Figure 3E). In addition, as demonstrated in Figure 3F, miR-195 was upregulated by TTN-AS1 knockdown in ACHN cells, as well as downregulated by TTN-AS1 overexpression in 786-O cells.
Cyclin D1 Is Directly Targeted by miR-195 in ccRCC
Then we tried to predict the possible target gene of miR-195 in ccRCC. By using the TargetScan database (http://www.targetscan.org/vert_71/), we noticed that the 3′-UTR region of cyclin D1 mRNA has the potential binding sites of miR-195 (Figure 4A). Dual-luciferase reporter assay showed that administration of miR-195 mimics significantly reduced the luciferase activity of cyclin D1-WT in both ACHN and 786-O cells (Figure 4B).
miR-195 Restoration Reverses the Effects of TTN-AS1 in ccRCC Cells
Rescue experiments further showed that the enhanced cell cycle progression in TTN-AS1-overexpressing 786-O cells was obviously blocked by co-transfection with miR-195 mimics (Figure 5A), accompanied by the reduction of cyclin D1 protein level (Figure 5B). MTT assay showed that restoration of miR-195 also abolished the enhanced proliferation of TTN-AS1-overexpressing 786-O cells (Figure 5C).
Discussion
ccRCC is an aggressive malignancy with a poor prognosis, and lots of lncRNAs have been found to serve a critical regulatory role in ccRCC progression. They may act as tumor suppressors or oncogenes. For example, it is reported that downregulation of lncRNA PGM5-AS1 correlates with tumor progression and predicts poor prognosis in ccRCC.11 Besides, He et al showed that lncRNA DLEU1 accelerates the malignant progression of ccRCC.12 Therefore, identification of ccRCC-related lncRNAs may help in the discovery of effective therapeutic strategies.
This research systematically investigated the potential role of lncRNA TTN-AS1 in ccRCC, and provided first evidence that TTN-AS1 was upregulated in ccRCC tissues and cell lines. Through a series of functional experiments, we further observed the effects of TTN-AS1 on the biological behaviors of ccRCC cells, showing that TTN-AS1 knockdown suppressed ccRCC cell proliferation partly by inducing cell cycle arrest, a hallmark of restrained cell proliferation.13 cyclin D1 has been well characterized as a crucial regulator of G1/S transition,14 and herein, the positive regulation of TTN-AS1 on cyclin D1 expression in ccRCC cells was also confirmed. These findings indicated that TTN-AS1 might serve as an oncogene in ccRCC.
Recently, the ceRNA hypothesis has attracted growing attention. It suggests that lncRNAs may act as competing endogenous RNAs (ceRNAs) in modulating the expression pattern and biological role of miRNAs.15 This regulation mode was widely confirmed in cancer biology.16 miR-195 is shown to be downregulated in a variety of human cancers, including ccRCC,17 and in this study, we also identified the direct binding relation between TTN-AS1 and miR-195 in ccRCC. Moreover, rescue experiments showed that restoration of miR-195 expression blocked the oncogenic effects of TTN-AS1 in ccRCC cells. miRNAs post-transcriptionally silence specific genes by binding to the target mRNAs. Researchers have reported that miR-195 directly targets cyclin D1 in osteosarcoma and cervical cancer,18,19 and herein, this interaction was also confirmed in ccRCC.
In conclusion, we confirmed that TTN-AS1 is overexpressed in ccRCC samples, and serves as an oncogenic lncRNA in ccRCC partly by working as a ceRNA of miR-195, and positively regulates cyclin D1 expression in ccRCC cells. This study reveals that a novel axis of TTN-AS1/miR-195/cyclin D1 is involved in ccRCC development, which might provide potential therapeutic targets for ccRCC patients.
Data Sharing Statement
All data generated or analyzed during this study are included in this article.
Disclosure
The authors declare that they have no competing interests.
References
1. Cheville JC, Lohse CM, Zincke H, Weaver AL, Blute ML. Comparisons of outcome and prognostic features among histologic subtypes of renal cell carcinoma. Am J Surg Pathol. 2003;27(5):612–624. doi:10.1097/00000478-200305000-00005
2. Gul A, Rini BI. Adjuvant therapy in renal cell carcinoma. Cancer. 2019;125(17):2935–2944. doi:10.1002/cncr.32144
3. Mattick JS, Makunin IV. Non-coding RNA. Hum Mol Genet. 2006;15(Spec No 1):R17–R29. doi:10.1093/hmg/ddl046
4. Miranda-Castro R, de-Los-Santos-Alvarez N, Lobo-Castanon MJ. Long noncoding RNAs: from genomic junk to rising stars in the early detection of cancer. Anal Bioanal Chem. 2019;411(19):4265–4275. doi:10.1007/s00216-019-01607-6
5. Wapinski O, Chang HY. Long noncoding RNAs and human disease. Trends Cell Biol. 2011;21(6):354–361. doi:10.1016/j.tcb.2011.04.001
6. Dong MM, Peng SJ, Yuan YN, Luo HP. LncRNA TTN-AS1 contributes to gastric cancer progression by acting as a competing endogenous RNA of miR-376b-3p. Neoplasma. 2019;66(04):564–575. doi:10.4149/neo_2018_180927N721
7. Zhong Y, Wang J, Lv W, Xu J, Mei S, Shan A. LncRNA TTN-AS1 drives invasion and migration of lung adenocarcinoma cells via modulation of miR-4677-3p/ZEB1 axis. J Cell Biochem. 2019;120(10):17131–17141.
8. Wang Y, Jiang F, Xiong Y, Cheng X, Qiu Z, Song R. LncRNA TTN-AS1 sponges miR-376a-3p to promote colorectal cancer progression via upregulating KLF15. Life Sci. 2019;244:116936. doi:10.1016/j.lfs.2019.116936
9. Cui Z, Han B, Wang X, Li Z, Wang J, Lv Y. Long non-coding RNA TTN-AS1 promotes the proliferation and invasion of colorectal cancer cells by activating miR-497-mediated PI3K/Akt/mTOR signaling. Onco Targets Ther. 2019;12:11531–11539. doi:10.2147/OTT.S229104
10. Livak KJ, Schmittgen TD. Analysis of relative gene expression data using real-time quantitative PCR and the 2(-Delta Delta C(T)) method. Methods. 2001;25(4):402–408. doi:10.1006/meth.2001.1262
11. Qian M, Zheng JL, Kang N, Su YL. Down-regulation of long noncoding RNA PGM5-AS1 correlates with tumor progression and predicts poor prognosis in clear cell renal cell carcinoma. Eur Rev Med Pharmacol Sci. 2019;23(24):10685–10690. doi:10.26355/eurrev_201912_19767
12. He GZ, Yu SY, Zhou QP, et al. LncRNA DLEU1 accelerates the malignant progression of clear cell renal cell carcinoma via regulating miRNA-194-5p. Eur Rev Med Pharmacol Sci. 2019;23(24):10691–10698. doi:10.26355/eurrev_201912_19768
13. Sherr CJ. Cancer cell cycles. Science. 1996;274(5293):1672–1677. doi:10.1126/science.274.5293.1672
14. Coqueret O. Linking cyclins to transcriptional control. Gene. 2002;299(1–2):35–55. doi:10.1016/S0378-1119(02)01055-7
15. Tay Y, Rinn J, Pandolfi PP. The multilayered complexity of ceRNA crosstalk and competition. Nature. 2014;505(7483):344–352. doi:10.1038/nature12986
16. Yang C, Wu D, Gao L, et al. Competing endogenous RNA networks in human cancer: hypothesis, validation, and perspectives. Oncotarget. 2016;7(12):13479–13490. doi:10.18632/oncotarget.7266
17. Sun P, Wang L, Lu Y, et al. MicroRNA-195 targets VEGFR2 and has a tumor suppressive role in ACHN cells via PI3K/Akt and Raf/MEK/ERK signaling pathways. Int J Oncol. 2016;49(3):1155–1163. doi:10.3892/ijo.2016.3608
18. Han K, Chen X, Bian N, et al. MicroRNA profiling identifies MiR-195 suppresses osteosarcoma cell metastasis by targeting CCND1. Oncotarget. 2015;6(11):8875–8889. doi:10.18632/oncotarget.3560
19. Li Z, Wang H, Wang Z, Cai H. MiR-195 inhibits the proliferation of human cervical cancer cells by directly targeting cyclin D1. Tumour Biol. 2016;37(5):6457–6463. doi:10.1007/s13277-015-4540-6
 © 2020 The Author(s). This work is published and licensed by Dove Medical Press Limited. The full terms of this license are available at https://www.dovepress.com/terms.php and incorporate the Creative Commons Attribution - Non Commercial (unported, v3.0) License.
By accessing the work you hereby accept the Terms. Non-commercial uses of the work are permitted without any further permission from Dove Medical Press Limited, provided the work is properly attributed. For permission for commercial use of this work, please see paragraphs 4.2 and 5 of our Terms.
© 2020 The Author(s). This work is published and licensed by Dove Medical Press Limited. The full terms of this license are available at https://www.dovepress.com/terms.php and incorporate the Creative Commons Attribution - Non Commercial (unported, v3.0) License.
By accessing the work you hereby accept the Terms. Non-commercial uses of the work are permitted without any further permission from Dove Medical Press Limited, provided the work is properly attributed. For permission for commercial use of this work, please see paragraphs 4.2 and 5 of our Terms.