Back to Journals » Cancer Management and Research » Volume 12
Long Non-Coding RNA OIP5-AS1 Contributes to Gallbladder Cancer Cell Invasion and Migration by miR-143-3p Suppression
Received 25 August 2020
Accepted for publication 26 November 2020
Published 17 December 2020 Volume 2020:12 Pages 12983—12992
DOI https://doi.org/10.2147/CMAR.S278719
Checked for plagiarism Yes
Review by Single anonymous peer review
Peer reviewer comments 3
Editor who approved publication: Dr Beicheng Sun
Jing Li, Hui Zhang, Hongwu Luo
Department of Hepatopancreatobiliary Surgery, The Third Xiangya Hospital, Central South University, Changsha 410013, Hunan, People’s Republic of China
Correspondence: Hongwu Luo
Department of Hepatopancreatobiliary Surgery, The Third Xiangya Hospital, Central South University, No. 138, Tongzipo Road, Yuelu District, Changsha, Hunan 410013, People’s Republic of China
Tel +86-15116115700
Email [email protected]
Objective: This study was designed to investigate the effect of long non-coding RNA (lncRNA) OIP5-AS1 on cell migration and invasion of gallbladder cancer (GBC) and its specific mechanism.
Methods: The expressions of lncRNA OIP5-AS1 and miR-143-3p in GBC cell lines (GBC-SD, SGC996 and NOZ) and gallbladder epithelial cells (HGBE cells) were measured by qRT-PCR. After loss- and gain-of-function experiments for OIP5-AS1 and miR-143-3p in GBC-SD cells, CCK-8 was applied to examine cell viability, cell scratch assay to measure cell migration, and transwell chamber to inspect cell invasion capacity. The interaction between OIP5-AS1 and miR-143-3p was predicted by StarBase. Then, luciferase reporter gene assay and RNA pull-down were used to verify the targeting relationship between miR-143-3p and OIP5-AS1.
Results: OIP5-AS1 was highly expressed and miR-143-3p was downregulated in GBC cell lines, when compared with HGBE cells. Overexpression of OIP5-AS1 or downregulation of miR-143-3p facilitated GBC-SD cell invasion, proliferation and migration, while different expression patterns were found in GBC-SD cells in response to OIP5-AS1 suppression or miR-143-3p overexpression. OIP5-AS1 negatively mediated miR-143-3p. MiR-143-3p upregulation partially reversed the inhibitory effect of OIP5-AS1 knockdown on GBC-SD cell activities.
Conclusion: LncRNA OIP5-AS1 accelerates the progression of GBC by suppressing miR-143-3p.
Keywords: OIP5-AS1, miR-143-3p, gallbladder cancer, proliferation, migration, invasion
Introduction
Gallbladder cancer (GBC), with high risk of distant metastasis and lymph node metastasis at the initial diagnosis, has emerged as one of the most prevalent gastrointestinal cancers.1 The poor prognosis of GBC may attribute to its aggressiveness and absence of symptoms in the early stages.2 Surgical resection is the only possible curative treatment, but the success rate is variable, depending on the stage of GBC and completeness of resection.3 Therefore, understanding the molecular changes related to GBC is of great significance for the development of effective therapy and early diagnosis, thereby improving the prognosis of GBC patients.
Long non-coding RNAs (lncRNAs), which act as genomically transcribed non-coding transcripts over 200 nucleotides, confer momentous impact on the deterioration or remission of cancer.4 In recent years, the coordinating roles of lncRNAs have been frequently reported in multiple biological processes of GBC progression, including migration, invasion and proliferation. For example, lncRNA LINC01694 interferes with the growth and invasion of GBC cells by regulating miR-340-5p to restrain Sox4.5 In GBC, GATA6-AS may restrain cancer cell invasion and migration through miR-421 knockdown.1 OPA-interacting protein 5 antisense RNA 1 (OIP5-AS1), which transcribed in the antisense of gene encoding OIP5, is implicated in the cancer progression.6 In laryngeal squamous cell carcinoma, OIP5-AS1 modulates migration and growth of cells.7 Additionally, OIP5-AS1 is responsible for oral squamous cell carcinoma regarding cell migration, proliferation and invasion through mediation of miR-338-3p/NRP1 axis.8 Nonetheless, the functional roles and precise expression patterns of OIP5-AS1 in GBC cells have not been clearly defined. MicroRNAs (miRNAs), performing themselves in GBC, are non-coding single-stranded RNAs that can bind to the 3ʹuntranslated regions of their target mRNAs to repress protein expression.9 Among miRNAs, miR-143-3p has been researched to suppress the growth and angiogenesis of GBC.10
Former studies proposed the possibility that lncRNA acts as a competing endogenous (ceRNA) for miRNA to regulate its targeting gene expression in the GBC.11,12 In consideration of the potential relationship of lncRNA and miRNA, we aimed to explore whether lncRNA OIP5-AS1 is engaged in the progression of GBC through targeting miR-143-3p in a ceRNA manner. This competitive role of lncRNA OIP5-AS1 is elicited to contribute to tumor progression. More evidence revealed that OIP5-AS1 promotes the progression of GBC through suppressing miR-143-3p.
Materials and Methods
Cell Culture
The Human GBC GBC-SD cell line was supplied by the Cell Bank of Chinese Academy of Sciences (Shanghai, China). SGC996 cell line was purchased from the Cancer Institute of Tongji University School of Medical (Shanghai, China). NOZ cell line was provided by Health Science Research Resources Bank (Osaka, Japan) and gallbladder epithelial cells (HGBE) were offered by ATCC (Gaithersburg, MD, USA). The GBC-SD, SGC996 and NOZ cell lines were soaked in 1640 culture medium (11879020, Gibco, New York, USA) supplemented with 10% fetal bovine serum (FBS, 16140071, Gibco, New York, USA), and the gallbladder epithelial cells (HGBE) were immersed in Dulbecco’s modified Eagle medium (DMEM, 11965092, Gibco, New York, USA) containing 10% FBS and 1% penicillin/streptomycin. Cell culture was conducted at 37°C gassed with 5% CO2. The following experiments were conducted when cell confluence reached 80% ~90%. The use of cell lines was approved by the Ethics Committee of the Third Xiangya Hospital of Central South University.
Cell Transfection and Grouping
The si-OIP5-AS1, si-NC, pcDNA3.1-OIP5-AS1, pcDNA3.1-NC, miR-143-3p antagomir, NC antagomir, miR-143-3p agomir and NC agomir were designed and synthesized by Invitrogen (Shanghai, China). Twenty-four h before transfection, cells were aspirated into a six-well plate (1 × 105 cells/well) with fresh serum-free medium. Following transfection based on the instruction of the Lipofectamine 3000 kit (L3000015, Invitrogen, New York, USA), cells were correspondingly assigned into the si-OIP5-AS1 group (2.5 μg), si-NC group (2.5 μg, negative control of si-OIP5-AS1), pcDNA3.1-OIP5-AS1group (2.5 μg), pcDNA3.1-NC group (2.5 μg, negative control of pcDNA3.1-OIP5-AS1), miR-143-3p agomir group (200 nmol), NC agomir (200 nmol, negative control of miR-143-3p agomir), miR-143-3p antagomir group (200 nmol), NC antagomir (200 nmol, negative control of miR-143-3p antagomir), si-OIP5-AS1+miR-143-3p antagomir group, si-NC+NC antagomir group, si-OIP5-AS1+NC antagomir group, si-NC+NC antagomir group, si-NC+miR-143-3p antagomir group and si-NC+NC antagomir group. Blank group was set as the negative control (exposure to the Lipofectamine 3000 kit without any plasmid). Measurement or detection in each group was performed 48 h after cell transfection.
RNA Isolation and qRT-PCR
GBC cells were dissolved in 1 mL of TRIzol (Thermo Fisher Scientific, MA, USA) and the total RNA was extracted. The cDNA template was obtained by reverse transcription of total RNA by using M-MLV reverse transcriptase and random primers. The reaction system was configured according to the instructions of Premix Ex TaqTMII kit (Takara, Dalian, China), the reaction conditions corresponding to each primer were set and the ABI7500 quantitative PCR instrument (Applied Biosystems, Shanghai, China) was employed for RT-PCR experiments. The internal reference was presented as GAPDH, and data analysis utilized 2−ΔΔCt method.13 The formula is as follows: ΔΔCt = [Ct(target gene)-Ct(reference gene)] experimental group-[Ct(target gene)-Ct(reference gene)] control group. The primer sequences are shown in Table 1.
 |
Table 1 Primer Sequence Information |
CCK-8 Assay
Cell proliferation rate was evaluated by using the CCK-8 kit (Beyotime, Shanghai, China). The transfected cells were seeded into 96-well plates (3000 cells/well), and the fresh medium was replaced every day. The proliferation of cells was assessed with the CCK-8 kit every 24 h for 5 consecutive days: 10 μL of CCK-8 solution was added to cells/well for 2 h of incubation at 37°C. The absorbance (optical density, OD) was assessed by SpectraMax M5 microplate reader (Molecular Devices) at 450 nm wavelength.
Cell Scratch Assay
Cells in the Control group and experimental group were aspirated into a 12-well plate (1 × 105 cells). Following reached 100% confluence, cells were scratched by using a 10 µL pipette tip. Dulbecco’s phosphate-buffered saline (DPBS, 14190250, Gibco, New York, USA) was used to wash the cells thrice to remove the scratched cells, and then cells were cultured in fresh DMEM covering 2% FBS. Photographs of cells in the same field of vision were captured under an inverted microscope (Olympus) at time 0 and 24 h post-cell scratch, and the change of scratch width was observed. The migrate rate = (scratch distance at 0 h – scratch distance at 24 h)/scratch distance at 0 h. The test was done in triplicate.
Transwell Assay
The invasion ability of cells was displayed by using a transwell chamber (Corning, New York, USA). Following 2 h of Matrigel coating at 37°C, the membrane was inserted in the middle of the two-chamber. Cells (3 × 104) suspended in culture medium with 1% FBS were pipetted into an apical chamber, and 0.8 mL medium with 10% FBS was added into the basolateral chamber. After 24 h of incubation, non-invaded cells on the upper compartment of the membrane were removed by using a cotton swab. The invaded cells on the bottom surface of the membrane were fixed in methyl alcohol for 30 min, followed by staining with 10% Giemsa and three times of PBS wash. Cell counting of invaded cells was done by using an inverted microscope, and three independent experiments were conducted to confirm the reproducibility.
Luciferase Reporter Gene Assay
The binding site of OIP5-AS1 and miR-143-3p was predicted by online prediction software TargetScan. The mutated type sequences and wild-type sequences (MT-OIP5-AS1, WT-OIP5-AS1) were accordingly designed, cloned and combined with pGL3-Basic (Promega, USA). After cotransfection of GBC-SD cells with MT-OIP5-AS1/WT-OIP5-AS1 and miR-143-3p agomir/miR-143-3p antagomir for 48 h, the luciferase activity was evaluated by the luciferase kit (RG051S, Beyotime).
RNA Pull-Down Assay
The RNA pull-down assay was conducted with the instruction of an RNA pull-down kit (20164, Thermo Fisher). In brief, OIP5-AS1 purified in vitro was transcribed by Takara kit (Takara, Dalian, China). Then, OIP5-AS1 was labeled with biotin using Pierce RNA 3ʹDesthiobiotinylation kit (20163, Thermo Fisher). To ensure the formation of the secondary structure of RNA, the synthesized RNA was heated at 95°C for 2 min, placed on ice for 2 min and then put at room temperature for 20 min. The streptavidin magnetic beads were incubated with biotin-labeled OIP5-AS1 to obtain molecules that interacted with OIP5-AS1. Then, the RNA-binding protein was harvested after folded magnetic bead-biotin labeled-OIP5-AS1 was stirred or rotated with cell lysate in RIP buffer at 4°C for 2 h. The beads were washed six times with RIP wash buffer, and the collected RNA was detected by qRT-PCR.
Statistical Analysis
Data were analyzed by employing SPSS 18.0 (IBM Corp., Armonk, NY, USA) and GraphPad Prism 6.0 (GraphPad Software Inc.). Data were displayed as mean ± standard deviation (SD). T test was applied to the comparison between two groups, whereas comparisons between multiple groups were conducted utilizing one-way analysis of variance (ANOVA). Significance was set at P < 0.05. All experiments were repeated three times unless otherwise specified.
Results
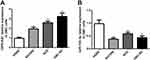
High Expression of OIP5-AS1 and Low Expression of miR-143-3p in GBC Cell Lines
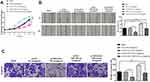
Result of qRT-PCR presented that compared with HGBE cells, the expression of OIP5-AS1 in GBC cell lines (GBC-SD cells, NOZ cells, SGC996 cells) was significantly increased (Figure 1A, P < 0.01), and miR-143-3p level was remarkably reduced in GBC cell lines (Figure 1B, P < 0.01). The maximum expression gap between OIP5-AS1 and miR-143-3p was found in GBC-SD cells, rather than the other two cell lines and therefore GBC-SD cells were most suitable for subsequent experiments.
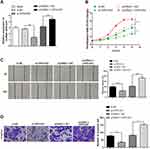
OIP5-AS1 Promotes Invasion, Proliferation and Migration of GBC Cells
To assess the role of lncRNA OIP5-AS1 in GBC-SD cells, GBC-SD cells were transfected with pcDNA3.1-OIP5-AS1 or si-OIP5-AS1. qRT-PCR results displayed good transfection efficiencies of pcDNA3.1-OIP5-AS1 and si-OIP5-AS1 in GBC-SD cells, as performed by elevated expression of OIP5-AS1 in pcDNA3.1-OIP5-AS1 group and decreased level of OIP5-AS1 in the si-OIP5-AS1 group (Figure 2A, P < 0.01). No obvious change was noticed in the si-NC, pcDNA3.1-NC group and Blank groups with regard to mRNA expression of OIP5-AS1 (Figure 2A, P > 0.05).
CCK-8 manifested that transfection with pcDNA3.1-OIP5-AS1 heightened GBC cell viability (Figure 2B, P < 0.01, vs the pcDNA3.1-NC group), while transfection with si-OIP5-AS1 repressed GBC cell viability (Figure 2B, P < 0.01, vs the si-NC group), suggesting that OIP5-AS1 can accelerate the growth of GBC-SD cells.
Results obtained from cell scratch assay and transwell revealed that overexpression of OIP5-AS1 enhanced the abilities of cell invasion (Figure 2D, P < 0.01) and migration (Figure 2C, P < 0.01), while knockdown of OIP5-AS1 diminished these abilities (Figure 2C and D, P < 0.01). These data illustrated that the biological functions of GBC cells can be enhanced by OIP5-AS1.
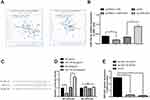
OIP5-AS1 Negatively Targets and Modulates miR-143-3p
The StarBase database was employed to predict the relationship between miRNAs and OIP5-AS1, and we found that miR-143-3p may be a potential target of OIP5-AS1. The correlation analysis by the online software StarBase2.0 showed the negative correlation between OIP5-AS1 and miR-143-3p in mesothelioma and adrenocortical cancer (Figure 3A). Based on this analysis, OIP5-AS1 was speculated to mediate the biological function of GBC-SD cells through miR-143-3p regulation.
Subsequently, qRT-PCR was applied to measure the expression level of miR-143-3p. The measurement showed increased mRNA level of miR-143-3p in the si-OIP5-AS1 group and decreased miR-143-3p expression in the pcDNA3.1-OIP5-AS1 group (Figure 3B, P < 0.01).
To verify the interaction between miR-143-3p and OIP5-AS1, the binding site of OIP5-AS1 and miR-143-3p was predicted, and the mutant and wide types of OIP5-AS1 and miR-143-3p were accordingly designed, which are depicted in Figure 3C. To ascertain targeting relationship between OIP5-AS1 and miR-143-3p, wild-type OIP5-AS1 luciferase promoter plasmid (WT-OIP5-AS1) and a mutant OIP5-AS1 luciferase promoter plasmid (MT-OIP5-AS1) were constructed. Result of luciferase reporter gene assay of the OIP5-AS1 promoter demonstrated that exposure to miR-143-3p agomir obviously reduced the luciferase activity of WT-OIP5-AS1 (Figure 3D, P < 0.01) while did not affect the luciferase activity of MT-OIP5-AS1. Additionally, after the RNA pull-down experiment and qRT-PCR detection, a large number of OIP5-AS1 were found in the bio-miR-143-3p-WT group (Figure 3E, P < 0.01), but there was no expression of OIP5-AS1 in the bio-miR-143-3p-MT group or bio-NC group (Figure 3E), indicating that miR-143-3p and OIP5-AS1 directly bound to each other. Together, there was a negative correlation of OIP5-AS1 and miR-143-3p in GBC cells.
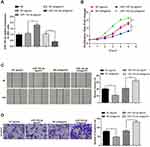
MiR-143-3p Inhibits Biological Functions of GBC Cells
The miR-143-3p was overexpressed or suppressed in GBC-SD cells to investigate the impact of miR-143-3p on GBC cells. Analysis of qRT-PCR displayed that the level of miR-143-3p was increased in the miR-143-3p agomir group (Figure 4A, P < 0.05, vs the NC agomir group) and suppressed in the miR-143-3p antagomir group (Figure 4A, P < 0.01, vs the NC antagomir group). No marked difference among the NC antagomir, NC agomir and Blank groups was noticed (Figure 4A, P > 0.05). These data manifested that miR-143-3p was notably repressed and overexpressed in GBC-SD cells, illustrating that miR-143-3p agomir and miR-143-3p antagomir with good transfection efficiencies can be applied to the subsequent experiments.
Then, the effect of miR-143-3p on the viability of GBC-SD cells was measured by the CCK-8 kit. We found that treatment with miR-143-3p antagomir increased cell viability (Figure 4B, P < 0.01, vs the NC antagomir group), while subjection to miR-143-3p agomir inhibited the ability of GBC cell proliferation (Figure 4B, P < 0.01, vs the NC agomir group).
The roles of miR-143-3p upregulation and miR-143-3p downregulation in the capacities of GBC-SD cell migration and invasion were assessed by cell scratch and transwell chamber experiments, respectively. The miR-143-3p antagomir group had enhanced abilities of cell invasion (Figure 4D, P < 0.01) and migration (Figure 4C, P < 0.01) in comparison to the NC antagomir group, while comparing with the NC agomir group, lower migration and invasion abilities were discovered in the miR-143-3p agomir group (Figure 4C and D, P < 0.01). Collectively, miR-143-3p can repress migration, invasion and growth of GBC cells.
OIP5-AS1 Governs Biological Functions of GBC Cells by miR-143-3p Downregulation
To investigate whether OIP5-AS1 contributes to the biological function of GBC-SD cells by inhibiting miR-143-3p, si-OIP5-AS1 and miR-143-3p antagomir were transfected or cotransfected into GBC-SD cells. Results of CCK-8 presented a pronounce increase in cell viability in the si-NC+miR-143-3p antagomir group, and a prominent decreased in the si-OIP5-AS1+NC antagomir group (Figure 5A, P < 0.01, vs the si-NC+NC antagomir group). Additionally, cell scratch assay showed that the si-NC+miR-143-3p antagomir group had elevated cell migration ability, while the si-OIP5-AS1+NC antagomir group possessed suppressed cell migratory property (Figure 5B, P < 0.01, vs the si-NC+NC antagomir group). The findings of the transwell assay revealed that there was higher cell invasion ability in the si-NC+miR-143-3p antagomir group, and lower cell invasive property in the si-OIP5-AS1+NC antagomir group (Figure 5C, P < 0.01, vs the si-NC+NC antagomir group). Compared with the si-NC+miR-143-3p antagomir group, the si-OIP5-AS1+miR-143-3p antagomir group possessed markedly weakened abilities of cell proliferation, migration and invasion (Figure 5A–C, P < 0.01). No notable difference was observed in the Blank, si-NC+NC antagomir and si-OIP5-AS1+miR-143-3p antagomir groups (Figure 5A–C, P > 0.05). These results revealed that miR-143-3p antagomir can eliminate the inhibitory effect of OIP5-AS1 knockdown on the GBC-SD cell activities. Taken these data together, OIP5-AS1 mediates miR-143-3p to regulate progression of GBC.
Discussion
GBC is a frequently diagnosed biliary tract with high incidence and mortality.14,15 Here, GBC-SD cells were selected from GBC cell lines and used for the subsequent experiments. We put the spotlight on the function of lncRNA OIP5-AS1 or miR-143-3p in GBC progression after identified the differentially expressed OIP5-AS1 and miR-143-3p in GBC-SD cells. Herein, we proposed that the invasion, migration and proliferation of GBC cells can be accelerated by the OIP5-AS1/miR-143-3p axis.
LncRNAs inactivate tumor suppressors or activate oncogenes to trigger the growth or metastasis phenotype of cancer cells by interacting with specific RNA-binding proteins.16 Herein, we characterized OIP5-AS1, which is transcribed in the antisense direction from the same gene that encodes OIP5.17 LncRNA OIP5-AS1 is reported to be upregulated and promotes cell proliferation in lung cancer.18 An important implication of our findings was that the high expression of OIP5-AS1 was identified in GBC-SD cells (Figure 1A, P < 0.01). Then, the loss- and gain-of-function experiments for lncRNA OIP5-AS1 were performed to explore whether overexpressed or silenced OIP5-AS1 affects the activities of GBC cells. Functionally, CCK-8, cell scratch and transwell assays showed that amplifying OIP5-AS1 in GBC cells heightened capacities of cell migration, invasion and proliferation, whereas knockdown of OIP5-AS1 had opposite results (Figure 2B–D, P < 0.01). Growing evidence proposed the oncogenic role of OIP5-AS1 in kinds of cancers. For instance, the growth, metastasis and EMT progress of hepatoblastoma are elicited by OIP5-AS1 through the miR-186a-5p/ZEB1 axis.19 OIP5-AS1 enhances CDK14 expression to induce osteosarcoma tumorigenesis by targeting miR-223.20 LncRNA OIP5-AS1 drives hemangioma endothelial cell invasion, proliferation and migration through the miR-195-5p/NOB1 axis modulation.21 These data proved that OIP5-AS1 is an oncogene to participate in GBC, and we speculated that the implication of OIP5-AS1 in GBC cells may be in a ceRNA manner.
The former study suggested that OIP5-AS1 performs itself in cervical cancer, microglioma, hemangioma and other tumors, and expressed in the cytoplasm through a ceRNA manner.22,23 MiR-143-3p is reported to be abnormally expressed and directly correlated with the prognosis and progression of various cancers.24,25 Of note, miR-143-5p dramatically represses the invasion, proliferation and migration of GBC cells through targeting the HIF-1α/EMT-related signaling pathway.26 Toward this end, the relationship between OIP5-AS1 and miR-143-3p in GBC cells needs to be clarified. Subsequently, the negative correlation between OIP5-AS1OIP5-AS1 and miR-143-3p in mesothelioma and adrenocortical cancer was exhibited by the online software StarBase2.0 (Figure 3A). Considering these findings, we speculated that lncRNA OIP5-AS1 interferes with the development of GBC through miR-143-3p regulation. Toward this end, RNA pull-down assay and luciferase reporter assay corroborated the binding relationship between miR-143-3p and OIP5-AS1 in GBC cells. In brief, OIP5-AS1 bound to miR-143-3p, and OIP5-AS1 negatively mediated miR-143-3p (Figure 3D and E). Subsequently, the results of CCK-8, cell scratch and transwell assays further confirmed that OIP5-AS1 may be responsible for cell migration, invasion and proliferation by regulating miR-143-3p in GBC (Figure 5A–C). This is supported by the recent work of Chen et al, who reported that the OIP5-AS1/miR-143-3p axis contributes to metastasis of cervical cancer.27 Additionally, these results are similar to those reported by Yang et al, which showed that lncRNA OIP5-AS1 accelerates proliferation and invasion through elevating integrin α6 expression by targeting miR-143-3p in cervical cancer.28
In summary, our findings identify the novel lncRNA OIP5-AS1 as an oncogene in GBC. The upregulation of OIP5-AS1 is associated with deterioration of GBC. Molecular mechanism exploration manifests that OIP5-AS1 enhances the biological activities of GBC cells regarding cell invasion, migration and proliferation by targeting miR-143-3p to accelerate GBC tumorigenesis. This study provides novel insights into the pathogenesis of GBC and potential therapeutic targets for this malignancy. Future targeted therapies likely offer the best hope for improving survival in GBC patients. However, there are some limitations in this study. First, the experimental data is limited. Second, no animal experiments and clinical experiments are done to further support our findings.
Acknowledgment
Thanks for the support from the Research of selenium-containing compound in suppressing CCL4-induced liver fibrosis in rats (KY040050).
Disclosure
The authors declare they have no conflicts of interest.
References
1. Li K, Tang J, Hou Y. LncRNA GATA6-AS inhibits cancer cell migration and invasion in gallbladder cancer by downregulating miR-421. Onco Targets Ther. 2019;12:8047–8053. doi:10.2147/OTT.S212231
2. Maplanka C. Gallbladder cancer, treatment failure and relapses: the peritoneum in gallbladder cancer. J Gastrointest Cancer. 2014;45(3):245–255. doi:10.1007/s12029-014-9597-8
3. Hickman L, Contreras C. Gallbladder cancer: diagnosis, surgical management, and adjuvant therapies. Surg Clin North Am. 2019;99(2):337–355. doi:10.1016/j.suc.2018.12.008
4. Wang SH, Zhang MD, Wu XC, Weng MZ, Zhou D, Quan ZW. Overexpression of LncRNA-ROR predicts a poor outcome in gallbladder cancer patients and promotes the tumor cells proliferation, migration, and invasion. Tumour Biol. 2016;37(9):12867–12875. doi:10.1007/s13277-016-5210-z
5. Liu L, Yan Y, Zhang G, Chen C, Shen W, Xing P. Knockdown of LINC01694 inhibits growth of gallbladder cancer cells via miR-340-5p/Sox4. Biosci Rep. 2020;40(4).
6. Meng X, Ma J, Wang B, Wu X, Liu Z. Long non-coding RNA OIP5-AS1 promotes pancreatic cancer cell growth through sponging miR-342-3p via AKT/ERK signaling pathway. J Physiol Biochem. 2020;76(2):301–315. doi:10.1007/s13105-020-00734-4
7. Wang H, Qian J, Xia X, Ye B. Long non-coding RNA OIP5-AS1 serves as an oncogene in laryngeal squamous cell carcinoma by regulating miR-204-5p/ZEB1 axis. Naunyn Schmiedebergs Arch Pharmacol. 2020;393(11):2177–2184. doi:10.1007/s00210-020-01811-7
8. Li M, Ning J, Li Z, et al. Long noncoding RNA OIP5-AS1 promotes the progression of oral squamous cell carcinoma via regulating miR-338-3p/NRP1 axis. Biomed Pharmacother. 2019;118:109259. doi:10.1016/j.biopha.2019.109259
9. Yang F, Tang Z, Duan A, et al. Long noncoding RNA NEAT1 upregulates survivin and facilitates gallbladder cancer progression by sponging microRNA-335. Onco Targets Ther. 2020;13:2357–2367. doi:10.2147/OTT.S236350
10. Jin YP, Hu YP, Wu XS, et al. miR-143-3p targeting of ITGA6 suppresses tumour growth and angiogenesis by downregulating PLGF expression via the PI3K/AKT pathway in gallbladder carcinoma. Cell Death Dis. 2018;9(2):182. doi:10.1038/s41419-017-0258-2
11. Hu YP, Jin YP, Wu XS, et al. LncRNA-HGBC stabilized by HuR promotes gallbladder cancer progression by regulating miR-502-3p/SET/AKT axis. Mol Cancer. 2019;18(1):167. doi:10.1186/s12943-019-1097-9
12. Wu XS, Wang F, Li HF, et al. LncRNA-PAGBC acts as a microRNA sponge and promotes gallbladder tumorigenesis. EMBO Rep. 2017;18(10):1837–1853. doi:10.15252/embr.201744147
13. Burja B, Kuret T, Janko T, et al. Olive leaf extract attenuates inflammatory activation and DNA damage in human arterial endothelial cells. Front Cardiovasc Med. 2019;6:56. doi:10.3389/fcvm.2019.00056
14. Mehrotra R, Tulsyan S, Hussain S, et al. Genetic landscape of gallbladder cancer: global overview. Mutat Res. 2018;778:61–71. doi:10.1016/j.mrrev.2018.08.003
15. Wu TJ, Xu B, Zhao GH, Luo J, Luo C. IL-37 suppresses migration and invasion of gallbladder cancer cells through inhibition of HIF-1alpha induced epithelial-mesenchymal transition. Eur Rev Med Pharmacol Sci. 2018;22(23):8179–8185.
16. Cai Q, Jin L, Wang S, et al. Long non-coding RNA UCA1 promotes gallbladder cancer progression by epigenetically repressing p21 and E-cadherin expression. Oncotarget. 2017;8(29):47957–47968. doi:10.18632/oncotarget.18204
17. Kim J, Noh JH, Lee SK, et al. LncRNA OIP5-AS1/cyrano suppresses GAK expression to control mitosis. Oncotarget. 2017;8(30):49409–49420. doi:10.18632/oncotarget.17219
18. Wang M, Sun X, Yang Y, Jiao W. Long non-coding RNA OIP5-AS1 promotes proliferation of lung cancer cells and leads to poor prognosis by targeting miR-378a-3p. Thorac Cancer. 2018;9(8):939–949. doi:10.1111/1759-7714.12767
19. Zhang Z, Liu F, Yang F, Liu Y. Kockdown of OIP5-AS1 expression inhibits proliferation, metastasis and EMT progress in hepatoblastoma cells through up-regulating miR-186a-5p and down-regulating ZEB1. Biomed Pharmacother. 2018;101:14–23. doi:10.1016/j.biopha.2018.02.026
20. Dai J, Xu L, Hu X, et al. Long noncoding RNA OIP5-AS1 accelerates CDK14 expression to promote osteosarcoma tumorigenesis via targeting miR-223. Biomed Pharmacother. 2018;106:1441–1447. doi:10.1016/j.biopha.2018.07.109
21. Zhang J, Zhao T, Tian L, Li Y. LncRNA OIP5-AS1 promotes the proliferation of hemangioma vascular endothelial cells via regulating miR-195-5p/NOB1 axis. Front Pharmacol. 2019;10:449. doi:10.3389/fphar.2019.00449
22. Liu X, Zheng J, Xue Y, et al. PIWIL3/OIP5-AS1/miR-367-3p/CEBPA feedback loop regulates the biological behavior of glioma cells. Theranostics. 2018;8(4):1084–1105. doi:10.7150/thno.21740
23. Ren X, He J, Qi L, et al. Prognostic and clinicopathologic significance of long non-coding RNA opa-interacting protein 5-antisense RNA 1 in multiple human cancers. Artif Cells Nanomed Biotechnol. 2020;48(1):353–361. doi:10.1080/21691401.2019.1709854
24. Shi H, Shen H, Xu J, Zhao S, Yao S, Jiang N. MiR-143-3p suppresses the progression of ovarian cancer. Am J Transl Res. 2018;10(3):866–874.
25. Wang F, Liu J, Zou Y, et al. MicroRNA-143-3p, up-regulated in H. pylori-positive gastric cancer, suppresses tumor growth, migration and invasion by directly targeting AKT2. Oncotarget. 2017;8(17):28711–28724. doi:10.18632/oncotarget.15646
26. He M, Zhan M, Chen W, et al. MiR-143-5p deficiency triggers EMT and metastasis by targeting HIF-1alpha in gallbladder cancer. Cell Physiol Biochem. 2017;42(5):2078–2092. doi:10.1159/000479903
27. Chen X, Xiong D, Yang H, et al. Long noncoding RNA OPA-interacting protein 5 antisense transcript 1 upregulated SMAD3 expression to contribute to metastasis of cervical cancer by sponging miR-143-3p. J Cell Physiol. 2019;234(4):5264–5275. doi:10.1002/jcp.27336
28. Yang J, Jiang B, Hai J, Duan S, Dong X, Chen C. Long noncoding RNA opa-interacting protein 5 antisense transcript 1 promotes proliferation and invasion through elevating integrin alpha6 expression by sponging miR-143-3p in cervical cancer. J Cell Biochem. 2019;120(1):907–916. doi:10.1002/jcb.27454
 © 2020 The Author(s). This work is published and licensed by Dove Medical Press Limited. The full terms of this license are available at https://www.dovepress.com/terms.php and incorporate the Creative Commons Attribution - Non Commercial (unported, v3.0) License.
By accessing the work you hereby accept the Terms. Non-commercial uses of the work are permitted without any further permission from Dove Medical Press Limited, provided the work is properly attributed. For permission for commercial use of this work, please see paragraphs 4.2 and 5 of our Terms.
© 2020 The Author(s). This work is published and licensed by Dove Medical Press Limited. The full terms of this license are available at https://www.dovepress.com/terms.php and incorporate the Creative Commons Attribution - Non Commercial (unported, v3.0) License.
By accessing the work you hereby accept the Terms. Non-commercial uses of the work are permitted without any further permission from Dove Medical Press Limited, provided the work is properly attributed. For permission for commercial use of this work, please see paragraphs 4.2 and 5 of our Terms.