Back to Journals » OncoTargets and Therapy » Volume 13
Long Non-Coding RNA-NEAT1 Promotes Cell Migration and Invasion via Regulating miR-124/NF-κB Pathway in Cervical Cancer
Authors Shen X , Zhao W, Zhang Y, Liang B
Received 21 June 2019
Accepted for publication 26 March 2020
Published 17 April 2020 Volume 2020:13 Pages 3265—3276
DOI https://doi.org/10.2147/OTT.S220306
Checked for plagiarism Yes
Review by Single anonymous peer review
Peer reviewer comments 2
Editor who approved publication: Dr Arseniy Yuzhalin
Xiaofang Shen,1,* Wei Zhao,1,* Yumei Zhang,2 Bin Liang2
1Department of Obstetrics-Gynecology, Dongying City People’s Hospital, Dongying City, Shandong Province 257091, People’s Republic of China; 2Department of Obstetrics-Gynecology, Dongying City Dongying District People’s Hospital, Dongying City, Shandong Province 257000, People’s Republic of China
*These authors contributed equally to this work
Correspondence: Xiaofang Shen
Department of Obstetrics-Gynecology, Dongying City People’s Hospital, No. 317, Nanyi Road, Dongying District, Dongying City, Shandong Province 257091, People’s Republic of China
Tel +86-13780757608
Email [email protected]
Background: This study aimed to investigate the regulatory role of lncRNA-NEAT1 on cervical cancer (CC) and the underlying molecular mechanisms.
Methods: The expression of lncRNA-NEAT1 and miR-124 was detected in CC tissues and cells (HeLa and SiHa cells) by qRT-RCR. The relation between lncRNA-NEAT1 expression and clinical parameters of CC patients was explored. The cell migration and invasion were detected by wound healing assay and transwell assay. The cell proliferation was detected by CCK-8 and anchorage-independent colony assay. The targeting relation between miR-124 and lncRNA-NEAT1 was predicted by TargetScan and identified by dual luciferase reporter gene and RNA pull-down assay. The expression of metastasis- (MMP-2 and MMP), EMT- (E-cadherin, N-cadherin and Vimentin), and NF-κB pathway-related factors (NF-κB p65, p-NF-κB p65 and IκBα) was detected by Western blot.
Results: The expression of lncRNA-NEAT1 was upregulated in CC tissues and cells and positively correlated with TNM stage and lymph node metastasis. Overexpression of lncRNA-NEAT1 promoted the proliferation, migration and invasion, influenced the expression of EMT markers, and activated NF-κB pathway in HeLa and SiHa cells. Silencing of lncRNA-NEAT1 exhibited opposite effects on HeLa and SiHa cells. LncRNA-NEAT1 could negatively regulate its target miR-124. MiR-124 reversed the effects of lncRNA-NEAT1 on the migration, invasion, EMT and NF-κB pathway of HeLa cells.
Conclusion: LncRNA-NEAT1 promoted the migration and invasion of CC cells via regulating miR-124/NF-κB pathway.
Keywords: cervical cancer, lncRNA-NEAT1, migration, invasion, miR-124/NF-κB pathway
Introduction
Cervical cancer (CC) is one of the frequently diagnosed cancers and the second most common malignancy in women all over the world.1 According to relevant statistics, there are nearly 530,000 new CC patients worldwide each year.2,3 Meanwhile, approximately 270,000 patients die due to CC each year.4 Despite great progresses have been made in the diagnostic techniques and therapeutic approaches, the long-term survival rate of CC patients remains poor due to recurrence and metastasis.5 Thus, it is urgent to find novel therapeutic targets and diagnostic biomarkers for CC.
Long non-coding RNAs (lncRNAs) are transcripts longer than 200 nucleotides that regulate gene expression at the transcriptional and posttranscriptional level.6,7 Emerging evidences have confirmed that lncRNAs play important roles in the occurrence and progression of malignancies, including CC.4 LncRNAs participate in the regulation of various cellular biological processes, including cell proliferation, cell cycle, migration, and invasion.8,9 For instance, lncRNA 00152 promotes cell proliferation in CC.10 Gao et al11 have indicated that lncRNA DANCR regulates cell proliferation and metastasis of CC. It is noteworthy that lncRNA nuclear-rich transcripts 1 (lncRNA-NEAT1) promotes the proliferation and invasion, and inhibits the apoptosis of CC cells.12 However, the potential regulatory mechanism of lncRNA-NEAT1 on CC still needs to be investigated.
MicroRNAs (miRNAs) are a class of small non-coding RNAs of 19–25 nucleotides in length that regulate gene expression in the post-transcriptional level.13 Accumulating evidence have indicated that miRNAs exert important regulatory effects in cellular biological processes of cancer, such as cell proliferation, apoptosis, migration, invasion, and cell cycle.14,15 Previous researches have indicated that miR-124 plays a vital role in cancer.16,17 LncRNAs can regulate the expression of target mRNAs by acting as molecular sponges for miRNAs, thereby influencing the development of tumor.18,19 Cheng et al20 have reported that lncRNA-NEAT1 promotes the progression of nasopharyngeal carcinoma by regulating miR-124/Nuclear factor-κB (NF-κB) pathway. However, the interaction between lncRNA-NEAT1 and miR-124 in CC remains unclear.
In this study, we investigated the regulatory effects of lncRNA-NEAT1 on the cellular biological processes of CC, as well as the molecular mechanisms involving miR-124/NF-κB pathway. Our results indicated that lncRNA-NEAT1 promoted the migration and invasion of CC cells via regulating miR-124/NF-κB pathway. Our findings may provide a new theoretical foundation for the treatment of CC.
Methods
Patients and Tissue Samples
A total of 72 patients with CC were screened from our hospital from January 2017 to December 2018. No patients had received chemotherapy or radiotherapy before surgery. The characteristics of CC patients are presented in Table 1. The tumor tissues and adjacent normal tissues were collected from CC patients by surgical resection. This study was approved by the Ethics Committee of Dongying City People’s Hospital, and was performed in accordance with the Declaration of Helsinki. All patients signed informed consent.
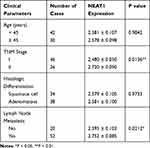 |
Table 1 Association Between LncRNA-NEAT1 Expression and Clinical Parameters of Patients with CC |
Cell Cultures
Normal human epidermal cell line HaCaT and human cervical cancer cell lines HeLa and SiHa were purchased from American Type Culture Collection (ATCC, USA). All the cells lines were cultured in Dulbecco’s Modified Eagle Medium (DMEM) (Gibco, USA) complemented with 10% fetal bovine serum (FBS, Gibco, USA) and 1% streptomycin/penicillin (Gibco, USA). All cells were cultured in a 5% CO2 incubator at 37°C.
Cell Transfection and Grouping
The full length sequence of NEAT1 was amplified by PCR and cloned into pcDNA3.1 vector (Invitrogen, USA) to construct pcDNA-NEAT1 overexpression plasmid (pcDNA-NEAT1). Empty pcDNA3.1 vector was considered as negative control (pcDNA-NC). The siRNA lncRNA-NEAT1 (si-NEAT1), siRNA negative control (si-NC), miR-124 mimics, and miR-124 negative control (miR-NC) were purchased from Ruibo (Guangzhou, China). HeLa and SiHa cells (1 × 105) were seeded into 6-well plates and cultured until 90% confluence. Cells were transfected with the above agents using Lipofectamine® 2000 Reagent (Invitrogen). HeLa cells were randomly divided into si-NEAT1, si-NC and BLANK group (no treatment). SiHa cells were randomly divided into pcDNA-NEAT1, pcDNA-NC and BLANK group (no treatment). In addition, HeLa cells were further divided into pcDNA-NC + miR-124 mimics, pcDNA-NC + miR-NC, pcDNA-NEAT1 + miR-124 mimics, and pcDNA-NEAT1 + miR-NC group. After 48 h of transfection, cells were used for subsequent experiments.
Wound Healing Assay
The transfected cells were inoculated into 6-well plates (1 × 105 cells/well) and cultured in a 5% CO2 incubator at 37°C. After 24 h of culturing, a wound track was scored with a 200 μL plastic scraper. The cell debris was removed by washing with PBS three times. The cells were then cultured in a 5% CO2 incubator at 37°C for 24 h. The scratch wound was observed and photographed under an optical microscope (100 ×).
Transwell Assay
The transfected cells at a density 1 × 105 cells/well were inoculated into the upper chamber. Then 600 μL medium with 10% FBS was added into the lower chamber. After 24 h of incubation at 37°C, cells on the lower chamber were fixed with 4% paraformaldehyde for 30 min and stained with 0.5% crystal violet (Sigma, USA) for 30 min at 25°C. For the detection of cell invasion, the upper chamber was per-coated with Matrigel (BD Biosciences, USA). The numbers of migrated and invaded cells were calculated under an optical microscope (200 ×).
CCK-8 Assay
The transfected cells were inoculated into 96-well plates (1 × 105 cells/well) and cultured in a 5% CO2 incubator at 37°C. At 24, 48, 72, and 96 h post-culturing, cells were incubated with CCK-8 solution (Invitrogen) for 4 h. The optical density (OD) at 450 nm was detected by a spectrophotometer (Bio-Rad, USA).
Anchorage-Independent Colony Assay
A base layer was formed in 6-well plates by using 0.7% agar-growth media solution. The transfected cells (1 × 104 cells/well) were gently mixed with 0.7% agar-media solution and then inoculated on the top of the base layer. After 2 weeks of incubation, the colonies were fixed with 4% paraformaldehyde for 10 min and stained with crystal violet for 20 min. The stained colonies were counted under an optical microscope (4 ×).
Dual Luciferase Reporter Gene Assay
TargetScan was used to predict the targeting relation between miR-124 and lncRNA-NEAT1. According to the binding sequences, lncRNA-NEAT1-Wt and lncRNA-NEAT1-Mut fragments were designed and cloned into pmirGLO luciferase vector (Promega, USA). HeLa and SiHa cells were co-transfected with miR-124 mimics/miR-124 mimics negative control (miR-NC) and lncRNA-NEAT1-Wt/lncRNA-NEAT1-Mut by Lipofectamine 2000 (Invitrogen, USA). After 48 h of transfection, the luciferase activity was detected by using a dual luciferase kit (Promega, USA).
RNA Pull-Down Assay
HeLa and SiHa cells were lysed using RIPA Lysis buffer (Sigma, USA), and incubated with biotin-labeled lncRNA-NEAT1 NC (Bio-NC), lncRNA-NEAT1-Wt (Bio-ncRNA-NEAT1-Wt) and lncRNA-NEAT1-Mut (ncRNA-NEAT1-Mut) for 1 h at 37°C. The samples were then incubated with Streptavidin agarose beads (Invitrogen) for 1 h at 37°C. The eluants were collected for detecting miR-124 expression by quantitative real-time PCR (qRT-PCR).
qRT-PCR
Total RNA was extracted from tissues and cells using TRIZOL agent (Invitrogen, USA). Total RNA was then reverse-transcribed into cDNA using the PrimeScript RT reagent Kit (TaKaRa, China). qRT-PCR was performed on a qRT-PCR instrument (Bio-Rad, USA) using SYBR Green PCR kit (TaKaRa, China). Primers used in this study were as follows: lncRNA-NEAT1 (forward): 5′-ATGCTTTCTCAACTTGTTGG-3′, (reverse): 5′-TCACCG CTCTTGGCCGTCACA-3′; GAPDH (forward): 5′-AGTAGTCACCTGTTGCTGG-3′, (reverse): 5′-TAATACGGAGACCTGTCTGGT-3′; miR-124 (forward): 5′-AGGCCUCUCUCUCCGUGUUCAC-3′, (reverse): 5′-CAGCCCCATTCTTGGCATTCAC-3′; U6 (forward): 5′-GCTTCGGCAGCACATATACTAAAAT-3′, (reverse): 5′-CGCTTCACGAATTTGCGTGTCAT-3′. GAPDH and U6 were used as internal controls. The relative expression level was calculated according to the 2−ΔΔCt method.
Western Blot
Total proteins were extracted from cells using RIPA lysis buffer (Sigma). The protein sample (50 μg) was separated by 10% sodium dodecyl sulfate-polyacrylamide gel electrophoresis (SDS-PAGE) and then transferred onto polyvinylidene difluoride membrane. After blocked with 5% non-fat milk for 2 h at room temperature, the membrane was incubated with the primary antibody (MMP-2, 1:1000, 4022s; MMP-9, 1:1000, 3852s; E-cadherin, 1:1000, #14472; N-cadherin, 1:1000, #13116; Vimentin, 1:1000, #5741; NF-κB p65, 1:1000, #8242; p- NF-κB p65, 1:1000, #3033; IκBα, 1;1000, #4814; GAPDH, 1:1000, #5174, Cell Signal, USA) overnight at 4°C. After three times of washing with TBST, the membrane was then incubated with horseradish peroxidase-labeled secondary antibody (anti-rabbit IgG, 1:5000, Sigma, USA) for 2 h at 25°C. The protein bands were visualized with an ECL system (Bio-Rad) and then quantified by Image Lab™ Software (Bio-Rad).
Immunofluorescence
HeLa and SiHa cells were fixed with 4% paraformaldehyde for 20 min at 4°C, and permeated with 0.1% Triton X-100 for 5 min. Cells were then blocked with 5% BSA for 30 min, and incubated with primary antibody (anti-NF-κB p65, 1:100, Abcam) overnight at 4°C. After three times of washing with PBS, cells were incubated with FITC-labeled secondary antibody (goat anti-rabbit IgG, 1:500, Abcam) for 2 h at 37°C. Followed by 10 min of staining with 4′, 6-diamidino-2-phenylindole (DAPI) at 25°C, the fluorescence was observed under a fluorescence microscope.
Statistical Analysis
All statistical analyses were performed using SPSS 22.0 Statistical Software (SPSS Inc., Chicago, IL). All experiments were performed in triplicate. Data were presented as mean ± standard deviation (SD). The differences among multi-groups were analyzed by the One-way ANOVA followed by the Tukey’s post hoc test. The differences between two groups were analyzed by the Student t-test. The correlation was analyzed by Spearman correlation analysis. P < 0.05 was considered to be statistically significant.
Results
The Expression of lncRNA-NEAT1 Is Upregulated in CC Tissues and Cells
qRT-PCR showed that the expression of lncRNA-NEAT1 was significantly increased in CC tissues compared with adjacent tissues (P < 0.001) (Figure 1A). The expression of lncRNA-NEAT1 in HeLa and SiHa cells was higher than that in HaCaT cells (P < 0.001) (Figure 1B). In addition, the expression of lncRNA-NEAT1 was positively correlated with tumor-node-metastasis (TNM) stage (P < 0.05) and lymph node metastasis (P < 0.05) in patients with CC. The expression of lncRNA-NEAT1 was not significantly associated with the age and histologic differentiation in patients with CC (P > 0.05) (Table 1).
LncRNA-NEAT1 Promotes the Migration and Invasion of HeLa and SiHa Cells
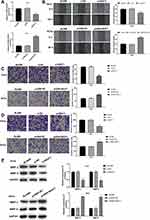
LncRNA-NEAT1 was overexpressed by the transfection of pcDNA-NEAT1, and silenced by the transfection of si-NEAT1 in HeLa and SiHa cells. As shown in Figure 2A, lncRNA-NEAT1 expression was significantly lower in the si-NEAT1 group, and was significantly higher in the pcDNA-NEAT1 group than that in the BLANK group (P < 0.001). Wound healing assay showed that the wound healing rate was significantly decreased in HeLa cells of the si-NEAT1 group, and was significantly increased in SiHa cells of the pcDNA-NEAT1 group compared with cells of the BLANK group (P < 0.001) (Figure 2B). In addition, transwell assay showed that the numbers of invaded and migrated cells were significantly decreased in HeLa cells of the si-NEAT1 group, and were significantly increased in SiHa cells of the pcDNA-NEAT1 group compared with cells of the BLANK group (P < 0.001) (Figure 2C and D). The expression of metastasis-related factors MMP-2 and MMP-9 was further detected in HeLa and SiHa cells. Western blot showed that the expression of MMP-2 and MMP-9 in HeLa cells was significantly decreased in the si-NEAT1 group compared with the BLANK group (P < 0.001). On the contrary, the expression of MMP-2 and MMP-9 in SiHa cells was significantly increased in the pcDNA-NEAT1 group compared with the BLANK group (P < 0.001) (Figure 2E). The migration and invasion of HeLa and SiHa cells were not significantly influenced by the transfection of si-NC and pcDNA-NC (Figure 2B–E). These results indicated that lncRNA-NEAT1 could promote the migration and invasion of HeLa and SiHa cells.
LncRNA-NEAT1 Promotes the Proliferation of HeLa and SiHa Cells
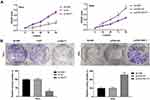
CCK-8 assay showed that the viability of HeLa cells in the si-NEAT1 group was significantly decreased compared with the BLANK group at 24, 48, 72 and 96 h post-culturing (P < 0.001). On the contrary, the viability of SiHa cells in the pcDNA-NEAT1 group was significantly increased compared with the BLANK group at different time points (P < 0.001) (Figure 3A). In addition, anchorage-independent colony assay showed that the number of colonies was significantly decreased in HeLa cells of the si-NEAT1 group, and was significantly increased in SiHa cells of the pcDNA-NEAT1 group compared with cells of the BLANK group (P < 0.01) (Figure 3B). The cell viability and colony formation of HeLa and SiHa cells were not significantly influenced by the transfection of si-NC and pcDNA-NC (Figure 3A and B). These result indicated that lncRNA-NEAT1 could promote the proliferation of HeLa and SiHa cells.
LncRNA-NEAT1 Influences the Expression of EMT Markers in HeLa and SiHa Cells
The expression of EMT-related factors E-cadherin, N-cadherin and Vimentin was detected in HeLa and SiHa cells. Western blot showed that the expression of E-cadherin in SiHa cells was significantly higher in the si-NEAT1 group than that in the BLANK group (P < 0.001). The expression of N-cadherin and Vimentin was significantly lower in the si-NEAT1 group than that in the BLANK group (P < 0.001) (Figure 4A). On the contrary, the expression of E-cadherin was significantly decreased, and the expression of N-cadherin and Vimentin was significantly increased in SiHa cells of the pcDNA-NEAT1 group compared with cells of the BLANK group (P < 0.001) (Figure 4B). The expression of E-cadherin, N-cadherin and Vimentin in HeLa and SiHa cells was not significantly influenced by the transfection of si-NC and pcDNA-NC (Figure 4A and B). These results suggested that lncRNA-NEAT1 could influence the EMT of HeLa and SiHa cells.
LncRNA-NEAT1 Activates NF-κB Pathway in HeLa and SiHa Cells
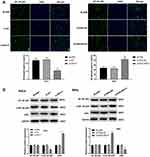
The nuclear expression of NF-κB p65 was detected by immunofluorescence. As shown in Figure 5A, the percentage of NF-κB p65-positive HeLa cells was significantly decreased in the si-NEAT1 group compared with the BLANK group (P < 0.01). The percentage of NF-κB p65-positive SiHa cells was significantly increased in the pcDNA-NEAT1 group compared with the BLANK group (P < 0.01). In addition, Western blot showed that the expression of p-NF-κB p65 was significantly decreased in HeLa cells of the si-NEAT1 group, and was significantly increased in SiHa cells of the pcDNA-NEAT1 group compared with cells of the BLANK group (P < 0.001). The expression of IκBα was opposite to that of p-NF-κB p65 in HeLa and SiHa cells. However, the expression of NF-κB p65 was not significantly changed in HeLa and SiHa cells by the transfection of si-NEAT1 and pcDNA-NEAT1 (Figure 5B). These results suggested that lncRNA-NEAT1 could activate NF-κB pathway in HeLa and SiHa cells.
LncRNA-NEAT1 Negatively Regulates the Expression of miR-124
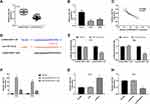
qRT-PCR showed that the expression of miR-124 was significantly lower in CC tissues than that in adjacent tissues (P < 0.001) (Figure 6A), and was significantly lower in HeLa and SiHa cells than that in HaCaT cells (P < 0.01) (Figure 6B). The expression of lncRNA-NEAT1 was negatively correlated with the expression of miR-124 in CC tissues (P < 0.001) (Figure 6C). A binding site of miR-124 at lncRNA-NEAT1 was predicated by Target Scan (Figure 6D). Dual luciferase reporter gene assay showed that the luciferase activity was significantly decreased in the miR-124 mimics + lncRNA-NEAT1-Wt group compared with the miR-NC + lncRNA-NC-Wt group (P < 0.001) (Figure 6E). RNA pull down assay showed that the expression of miR-124 was significantly increased in the Bio-lncRNA-NEAT1-Wt group compared with the Bio-NC group (P < 0.01) (Figure 6F). In addition, the transfection of si-NEAT1 significantly increased the expression of miR-124 in HeLa cells (P < 0.001) (Figure 6G). The transfection of pcDNA-NEAT1 significantly decreased the expression of miR-124 in SiHa cells (P < 0.001) (Figure 6H). These results indicated that lncRNA-NEAT1 could negatively regulate its target miR-124 in HeLa and SiHa cells.
MiR-124 Reverses the Effects of lncRNA-NEAT1 on the Migration, Invasion, EMT and NF-κB Pathway of HeLa Cells
To explore whether lncRNA-NEAT1 exerted its function in CC through regulating miR-124, HeLa cells were co-transfected with pcDNA-NEAT1 and miR-124 mimics. Wound healing and transwell assay showed that the migration and invasion ability of HeLa cells were decreased in the pcDNA-NC + miR-124 mimics group, and were increased in the pcDNA-NEAT1 + miR-NC group compared with the pcDNA-NC + miR-NC group (P < 0.001). The transfection of miR-124 mimics significantly reversed the promoting effects of pcDNA-NEAT1 on the migration and invasion ability of HeLa cells (P < 0.01) (Figure 7A). In addition, the expression of MMP-2, MMP-9, N-cadherin, Vimentin, and p-NF-κB p65 in HeLa cells was significantly decreased in the pcDNA-NC + miR-124 mimics group, and was increased in the pcDNA-NEAT1 + miR-NC group compared with the pcDNA-NC + miR-NC group (P < 0.001). The expression of E-cadherin and IκBα was opposite to that of MMP-2 (P < 0.001). The transfection of miR-124 mimics significantly reversed the regulatory effects of pcDNA-NEAT1 on the expression of the above proteins in HeLa cells (P < 0.01) (Figure 7B–D). These results indicated that miR-124 could reverse the effects of lncRNA-NEAT1 on the migration, invasion, EMT and NF-κB pathway in HeLa cells.
Discussion
CC is one of the most common gynecologic malignant tumors in the world. The main therapeutic methods for CC are surgical operation, chemotherapy and radiotherapy. The long-term survival rate of CC patients remains poor due to recurrence and metastasis.5 Hence, it is urgent to explore the novel molecular mechanism of CC and identify novel therapeutic targets. In this study, we demonstrated that lncRNA-NEAT1 could promote the proliferation, migration and invasion of CC cells by regulating miR-124/NF-κB pathway.
Recently, accumulating evidences have indicated that lncRNAs are involved in the development of malignancies, including CC.4 LncRNAs have been reported to be abnormally expressed in tumors. Guo et al have confirmed that the expression of lncRNA-NEAT1 is significantly upregulated in CC tissues compared with the adjacent tissues.12 In consistent with the previous study, we found that lncRNA-NEAT1 was overexpressed in CC tissues, and HeLa and SiHa cells. In addition, the expression of lncRNA-NEAT1 was positively correlated with TNM stage and lymph node metastasis in CC patients. Previous studies have proved that lncRNAs play key regulatory roles in the proliferation, apoptosis, migration and invasion of tumor cells.8,21 For example, lncRNA TDRG1 promotes cell proliferation, migration and invasion through sponging miR-326 to modulate MAPK1 expression in CC.22 Zhu et al23 have indicated that lncRNA TUSC8 suppresses the invasion and migration of CC cells by miR-641/PTEN axis. Liu et al24 have reported that lncRNA MNX1-AS1 promotes the proliferation and inhibits the apoptosis of CC cells via MAPK pathway. In this study, we found that lncRNA-NEAT1 promoted the proliferation, migration and invasion of CC cells. MMP-2 and MMP-9 are involved in the process of tumor metastasis by regulating the migration and invasion of cancer cells.25 Song et al26 have proved that MMP-2 and MMP-9 expression are significantly increased in CC. Here, our results showed that lncRNA-NEAT1 increased the expression of MMP-2 and MMP-9 in CC cells.
NF-κB is a dimeric transcriptional factor that can regulate multiple cellular biological processes, such as cell proliferation, apoptosis, migration and invasion.27 Under normal physiological condition, the inactive NF-κB is sequestered in the cytoplasm by the inhibitor of κB (IκB) proteins.28 Upon stimulation, activated IκB kinase complex (IKK) promotes the phosphorylation and subsequent degradation of IκBα.28 Subsequently, NF-κB translocates to the nucleus where it regulates targeted gene expression.29 In this study, lncRNA-NEAT1 promoted the nuclear transfer of NF-κB p65, increased the phosphorylation of NF-κB p65 and decreased IκBα expression in CC cells. These results indicate that lncRNA-NEAT1 activates NF-κB pathway in CC cells. In addition, NF-κB pathway is essential for the process of EMT in cancer cells.31 EMT is a cellular transition from an epithelial to a mesenchymal state, which can be regulated at multilayer levels, including transcriptional control of gene expression, regulation of RNA splicing, and translational/post-translational control.32 Distinct EMT transition states present different functions, with the hybrid EMT state presenting the highest metastatic potential.35 EMT plays a vital role in the metastasis of CC, which is a significant phenotypic change through the acquisition of mesenchymal marker proteins (N-cadherin and Vimentin) and the loss of epithelial marker proteins (E-cadherin).34 In this study, lncRNA-NEAT1 decreased E-cadherin expression and increased the expression of N-cadherin and Vimentin in CC cells. These results suggest that the overexpression of lncRNA-NEAT1 may promote the transition of CC cells from an epithelial to a mesenchymal state. The hybrid EMT state may contribute to the metastasis of CC.
LncRNA-NEAT1 functions as a ceRNA of miRNAs, which plays an important regulatory role in cancer by antagonizing miRNAs functions and modulating miRNAs targets.20 For example, lncRNA-NEAT1 promotes cell growth and invasion through miR-211/HMGA2 axis in breast cancer.36 Here, miR-124 was identified as a target of lncRNA-NEAT1. Overexpression of lncRNA-NEAT1 inhibited the expression of miR-124 in CC cells, suggesting that lncRNA-NEAT1 negatively regulated miR-124. We further explored the regulatory role of miR-124 in CC. The results showed that miR-124 inhibited the cell migration and invasion, and blocked NF-κB pathway in HeLa cells. Note worthily, miR-124 reversed the effects of lncRNA-NEAT1 on the migration, invasion, EMT and NF-κB pathway of HeLa cells. All these results suggested that lncRNA-NEAT1 may promote the migration and invasion of CC cells by targeting miR-124.
Conclusion
LncRNA-NEAT1 was upregulated in CC tissues and cells, and positively correlated with TNM stage and lymph node metastasis in CC patients. Overexpression of lncRNA-NEAT1 promoted the proliferation, migration and invasion, influenced the EMT, and activated NF-κB pathway in CC cells. LncRNA-NEAT1 promoted the migration and invasion of CC cells via regulating miR-124/NF-κB pathway. Our findings provide a novel regulatory mechanism of lncRNA-NEAT1 in CC.
Data Sharing Statement
The data that support the findings of this study are all included in this paper.
Disclosure
The authors declare that there is no conflict of interest regarding the publication of this article.
References
1. Tsikouras P, Zervoudis S, Manav B, et al. Cervical cancer: screening, diagnosis and staging. J BUON. 2016;21:320–325.
2. Mazdziarz A, Wygledowski J, Osuch B, et al. New directions in cervical cancer prophylaxis worldwide and in Poland – case study of the polish rural female population. Ann Agric Environ Med. 2017;24(4):592–595. doi:10.5604/12321966.1232093
3. Ronco G, Franceschi S. Cervical cancer screening: the transformational role of routine human papillomavirus testing. Ann Intern Med. 2018;168(1):75–76. doi:10.7326/M17-2872
4. Zhang YY, Li M, Xu YD, et al. LncRNA SNHG14 promotes the development of cervical cancer and predicts poor prognosis. Eur Rev Med Pharmacol Sci. 2019;23:3664–3671. doi:10.26355/eurrev_201905_17790
5. Yee GP, de Souza P, Khachigian LM. Current and potential treatments for cervical cancer. Curr Cancer Drug Targets. 2013;13(2):205–220. doi:10.2174/1568009611313020009
6. Taheri M, Omrani MD, Ghafouri-Fard S. Long non-coding RNA expression in bladder cancer. Biophys Rev. 2018;10(4):1205–1213. doi:10.1007/s12551-017-0379-y
7. Nikpayam E, Tasharrofi B, Sarrafzadeh S, et al. The role of long non-coding RNAs in ovarian cancer. Iran Biomed J. 2017;21(1):3–15. doi:10.18869/acadpub.ibj.21.1.3
8. Jiang C, Li X, Zhao H, et al. Long non-coding RNAs: potential new biomarkers for predicting tumor invasion and metastasis. Mol Cancer. 2016;15(1):62. doi:10.1186/s12943-016-0545-z
9. Chen L, Dzakah EE, Shan G. Targetable long non-coding RNAs in cancer treatments. Cancer Lett. 2018;418:119–124. doi:10.1016/j.canlet.2018.01.042
10. Zheng JJ, Du XJ, Wang HP, et al. Long non-coding RNA 00152 promotes cell proliferation in cervical cancer via regulating miR-216b-5p/HOXA1 axis. Eur Rev Med Pharmacol Sci. 2019;23:3654–3663. doi:10.26355/eurrev_201905_17789
11. Cao L, Jin H, Zheng Y, et al. DANCR-mediated microRNA-665 regulates proliferation and metastasis of cervical cancer through the ERK/SMAD pathway. Cancer Sci. 2019;110:913–925. doi:10.1111/cas.13921
12. Guo HM, Yang SH, Zhao SZ, et al. LncRNA NEAT1 regulates cervical carcinoma proliferation and invasion by targeting AKT/PI3K. Eur Rev Med Pharmacol Sci. 2018;22:4090–4097. doi:10.26355/eurrev_201807_15400
13. Bartel DP. Bartel DP MicroRNAs: genomics, biogenesis, mechanism, and function. Cell. 2004;116:281–297. doi:10.1016/s0092-8674(04)00045-5
14. Lin JF, Zeng H, Zhao JQ. MiR-212-5p regulates the proliferation and apoptosis of AML cells through targeting FZD5. Eur Rev Med Pharmacol Sci. 2018;22:8415–8422. doi:10.26355/eurrev_201812_16540
15. Liu L, Ren W, Chen K. MiR-34a promotes apoptosis and inhibits autophagy by targeting HMGB1 in acute myeloid leukemia cells. Cell Physiol Biochem. 2017;41(5):1981–1992. doi:10.1159/000475277
16. Liang YJ, Wang QY, Zhou CX, et al. MiR-124 targets slug to regulate epithelial-mesenchymal transition and metastasis of breast cancer. Carcinogenesis. 2013;34:713–722. doi:10.1093/carcin/bgs383
17. Wang X, Wu Q, Xu B, et al. MiR-124 exerts tumor suppressive functions on the cell proliferation, motility and angiogenesis of bladder cancer by fine-tuning UHRF1. FEBS J. 2015;282(22):4376–4388. doi:10.1111/febs.13502
18. Liz J, Esteller M. lncRNAs and microRNAs with a role in cancer development. Biochim Biophys Acta. 2016;1859(1):169–176. doi:10.1016/j.bbagrm.2015.06.015
19. Yoon JH, Abdelmohsen K, Gorospe M. Functional interactions among microRNAs and long noncoding RNAs. Semin Cell Dev Biol. 2014;34:9–14. doi:10.1016/j.semcdb.2014.05.015
20. Cheng N, Guo Y. Long noncoding RNA NEAT1 promotes nasopharyngeal carcinoma progression through regulation of miR-124/NF-kappaB pathway. Onco Targets Ther. 2017;10:5843–5853. doi:10.2147/OTT.S151800
21. Gao D, Lv AE, Li HP, et al. LncRNA MALAT-1 elevates HMGB1 to promote autophagy resulting in inhibition of tumor cell apoptosis in multiple myeloma. J Cell Biochem. 2017;118:3341–3348. doi:10.1002/jcb.25987
22. Jiang H, Liang M, Jiang Y, et al. The lncRNA TDRG1 promotes cell proliferation, migration and invasion by targeting miR-326 to regulate MAPK1 expression in cervical cancer. Cancer Cell Int. 2019;19(1):152. doi:10.1186/s12935-019-0872-4
23. Zhu Y, Liu B, Zhang P, et al. LncRNA TUSC8 inhibits the invasion and migration of cervical cancer cells via miR-641/PTEN axis. Cell Biol Int. 2019;43(7):781–788. doi:10.1002/cbin.11152
24. Liu X, Yang Q, Yan J, et al. LncRNA MNX1-AS1 promotes the progression of cervical cancer through activating MAPK pathway. J Cell Biochem. 2019;120(3):4268–4277. doi:10.1002/jcb.27712
25. Yan SP, Chu DX, Qiu HF, et al. LncRNA LINC01305 silencing inhibits cell epithelial-mesenchymal transition in cervical cancer by inhibiting TNXB-mediated PI3K/Akt signalling pathway. J Cell Mol Med. 2019;23:2656–2666. doi:10.1111/jcmm.14161
26. Song C, Zhu S, Wu C, et al. Histone deacetylase (HDAC) 10 suppresses cervical cancer metastasis through inhibition of matrix metalloproteinase (MMP) 2 and 9 expression. J Biol Chem. 2013;288(39):28021–28033. doi:10.1074/jbc.M113.498758
27. Escarcega RO, Fuentes-Alexandro S, Garcia-Carrasco M, et al. The transcription factor nuclear factor-kappa B and cancer. Clin Oncol (R Coll Radiol). 2007;19(2):154–161. doi:10.1016/j.clon.2006.11.013
28. Huang W, Cui X, Chen J, et al. Long non-coding RNA NKILA inhibits migration and invasion of tongue squamous cell carcinoma cells via suppressing epithelial-mesenchymal transition. Oncotarget. 2016;7(38):62520–62532. doi:10.18632/oncotarget.11528
29. Hayden MS, Ghosh S. Signaling to NF-kappaB. Genes Dev. 2004;18:2195–2224. doi:10.1101/gad.1228704
30. Karin M, Ben-Neriah Y. Phosphorylation meets ubiquitination: the control of NF-[kappa]B activity. Annu Rev Immunol. 2000;18:621–663. doi:10.1146/annurev.immunol.18.1.621
31. Wang Y, Lin Z, Sun L, et al. Akt/Ezrin Tyr353/NF-kappaB pathway regulates EGF-induced EMT and metastasis in tongue squamous cell carcinoma. Br J Cancer. 2014;110:695–705. doi:10.1038/bjc.2013.770
32. Simeone P, Trerotola M, Franck J, et al. The multiverse nature of epithelial to mesenchymal transition. Semin Cancer Biol. 2019;58:1–10. doi:10.1016/j.semcancer.2018.11.004
33. Wang T, Shi F, Wang J, et al. Kallistatin suppresses cell proliferation and invasion and promotes apoptosis in cervical cancer through blocking NF-kappaB signaling. Oncol Res. 2017;25:809–817. doi:10.3727/096504016X14799180778233
34. Fan MJ, Zou YH, He PJ, et al. Long non-coding RNA SPRY4-IT1 promotes epithelial-mesenchymal transition of cervical cancer by regulating the miR-101-3p/ZEB1 axis. Biosci Rep. 2019;39. doi:10.1042/BSR20181339
35. Pastushenko I, Blanpain C. EMT transition states during tumor progression and metastasis. Trends Cell Biol. 2019;29(3):212–226. doi:10.1016/j.tcb.2018.12.001
36. Li X, Wang S, Li Z, et al. The lncRNA NEAT1 facilitates cell growth and invasion via the miR-211/HMGA2 axis in breast cancer. Int J Biol Macromol. 2017;105:346–353. doi:10.1016/j.ijbiomac.2017.07.053
 © 2020 The Author(s). This work is published and licensed by Dove Medical Press Limited. The full terms of this license are available at https://www.dovepress.com/terms.php and incorporate the Creative Commons Attribution - Non Commercial (unported, v3.0) License.
By accessing the work you hereby accept the Terms. Non-commercial uses of the work are permitted without any further permission from Dove Medical Press Limited, provided the work is properly attributed. For permission for commercial use of this work, please see paragraphs 4.2 and 5 of our Terms.
© 2020 The Author(s). This work is published and licensed by Dove Medical Press Limited. The full terms of this license are available at https://www.dovepress.com/terms.php and incorporate the Creative Commons Attribution - Non Commercial (unported, v3.0) License.
By accessing the work you hereby accept the Terms. Non-commercial uses of the work are permitted without any further permission from Dove Medical Press Limited, provided the work is properly attributed. For permission for commercial use of this work, please see paragraphs 4.2 and 5 of our Terms.