Back to Journals » OncoTargets and Therapy » Volume 13
Long Non-Coding RNA NCK1-AS1 Serves an Oncogenic Role in Gastric Cancer by Regulating miR-137/NUP43 Axis
Authors Li W, Duan J, Shi W, Lei L, Lv P
Received 22 April 2020
Accepted for publication 7 July 2020
Published 6 October 2020 Volume 2020:13 Pages 9929—9939
DOI https://doi.org/10.2147/OTT.S259336
Checked for plagiarism Yes
Review by Single anonymous peer review
Peer reviewer comments 2
Editor who approved publication: Prof. Dr. Takuya Aoki
Wenxing Li, Jiming Duan, Wenbin Shi, Liqiang Lei, Pin Lv
Department of General Surgery, The Second Hospital of Shanxi Medical University, Taiyuan 030001, People’s Republic of China
Correspondence: Wenxing Li Department of General Surgery
The Second Hospital of Shanxi Medical University, No. 382 Wuyi Street, Taiyuan 030001, People’s Republic of China
Email [email protected]
Introduction: Long non-coding RNA (lncRNA) NCK1-AS1 could regulate multiple cancer progression. However, little is known regarding the roles and acting mechanisms of NCK-AS1 in gastric cancer (GC) progression. This work was aimed to explore the relationship between NCK1-AS1 and GC progression to illustrate the mechanisms of NCK1-AS1.
Methods: NCK1-AS1 expression level in GC tissues and cells was measured with a quantitative real-time PCR method. In vitro experiments including cell counting kit-8 assay, colony formation assay, wound-healing assay, and transwell invasion assay were employed to detect biological roles of NCK1-AS1 in GC progression. In vivo experiments were performed to analyze the roles of NCK1-AS1 on GC malignant phenotype. Moreover, mechanisms behind the biological roles of NCK1-AS1 in GC were investigated using bioinformatic analysis, luciferase activity reporter assay, RNA immunoprecipitation assay, and rescue experiments.
Results: NCK1-AS1 was found to have elevated expression in GC tissues and cells in comparison with normal counterparts. Loss-of-function experiments showed knockdown of NCK1-AS1 refrained GC cell proliferation, colony formation, migration, and invasion in vitro. Animal experiments showed silence of NCK1-AS1 suppresses tumor growth in vivo. Functionally, NCK1-AS1 serves as a sponge for microRNA-137 (miR-137) to upregulate nucleoporin 43 (NUP43) expression in GC. Rescue experiments proved the carcinogenic role of NCK1-AS1/miR-137/NUP43 axis in GC progression.
Discussion: In conclusion, the NCK1-AS1/miR-137/NUP43 axis was identified that could contribute to GC malignancy behaviors.
Keywords: gastric cancer, NCK1-AS1, miR-137, nucleoporin 43, tumor treatment
Introduction
The mortality rate of gastric cancer (GC) ranks 2nd place among all cancer types worldwide.1 The prognosis of GC remains very poor due to late diagnosis or cancer metastasis.2 Hence, it is necessary to explore the mechanisms associated with GC initiation and progression with the aim to provide biomarkers for cancer diagnosis or treatment.
Non-coding RNAs (ncRNAs) represent about 98–99% of all human genome transcripts.3 Long non-coding RNA (lncRNA) is a type of ncRNA with a length of over 200 nucleotides.4 Recent studies indicated lncRNAs can participate in almost all biological and pathological processes including cell growth, metastasis, and drug resistance.5,6 There are multiple lncRNAs that have been identified to be abnormally expressed in GC and play vital roles in affecting GC carcinogenesis.7,8 NCK1-AS1 is recently found to be aberrantly expressed in multiple cancers but its role in GC remains to be explored.9,10
MicroRNA (miRNA) is another type of ncRNA with a length of 18–25 nucleotides.11 miRNA can affect target gene expression through a 3ʹ-untranslated region binding manner. Multiple miRNAs have been found with altered expression in GC but only some of them were functionally characterized.12 More and more evidence in recent years implied that lncRNAs and protein-coding genes could serve as competitive endogenous RNAs (ceRNAs) to compete for miRNA recognition elements, and thus affect the cellular physiological status.13
In this work, we aimed to explore expression levels and involvements of NCK1-AS1 in regulating GC progression. Moreover, we construct a lncRNA-miRNA-mRNA regulatory network to explore mechanisms by which NCK1-AS1 regulates GC progression. Our work presented here was the first to unveil the roles of lncRNA NCK1-AS1 in GC and may provide a novel therapeutic target for GC.
Materials and Methods
Clinical Samples
The study protocol was approved by the ethic committee of The Second Hospital of Shanxi Medical University. Written informed consent was obtained from all participants. Fifty-two paired tumor tissues and normal tissues were obtained from patients who underwent surgical treatment at The Second Hospital of Shanxi Medical University. These patients did not receive anti-cancer treatments before enrolled in this study. These tissues were frozen by liquid nitrogen and stored at −80°C until further usage.
Cells
Human gastric epithelial cell line (GES-1) and GC cells (BGC-823 and MGC-803) were obtained from ATCC (Manassas, VA, USA). Dulbecco’s modified Eagle’s medium (DMEM) in supplement with 10% fetal bovine serum (FBS, both from Thermo Fisher Scientific, Inc., Waltham, MA, USA) was used to incubate these cells at 37°C in a moist incubator containing 5% CO2.
Cell Transfection
Small interfering RNA against NCK1-AS1 (si-NCK1-AS1) and negative control (si-NC) were designed by RiboBio (Guangzhou, China). The mimic for miR-137 (miR-137 mimic), an inhibitor for miR-137 (miR-137 inhibitor), and negative control (mi-NC) were purchased from GenePharma (Shanghai, China). Nucleoporin 43 (NUP43) overexpression plasmid (pc-NUP43) and empty vector pcDNA3.1 were obtained from Generay (Shanghai, China). Cells were grown on 6-well plates and transfected with oligonucleotides or plasmids using Lipofectamine 2000 (Thermo Fisher Scientific, Inc.) according to the manufacturer’s instructions. Cells were transfected for 48 h and collected for the following analyses.
RNA Extraction and Quantitative Reverse-Transcription PCR (RT-qPCR)
RNAs were extracted using TRIzol reagent (Thermo Fisher Scientific, Inc.) and quantified with NanoDrop 2000 (Thermo Fisher Scientific, Inc.). RNA was reverse transcribed into cDNA using PrimeScript RT Mix (Takara, Dalian, China). RT-qPCR was conducted with SYBR Green (Takara) with an ABI 7900 system (Applied Biosystems, Foster City, CA, USA). Primers were as follows: NCK1-AS1: 5ʹ-TTCCCATTTCTCCCAGGTCC-3ʹ (forward) and 5ʹ-TGGTTACTTTGAGCCTGCCT-3ʹ (reverse); NUP43: 5ʹ-GTCCTGAAATAGTGTCCGTGGGT-3ʹ (forward) and 5ʹ-TATGTGCTGCTGGTTTGGGTGT-3ʹ (reverse); GAPDH, 5′-GAGTCCACTGGCGTCTTC-3′ (forward) and 5′-GATGATCTTGAGGCTGTTGTC-3′ (reverse); miR-137: 5ʹ-TTATTGCTTAAGAATACGCG-3ʹ (forward) and 5ʹ-TCGTATCCAGTGCAGGGTC-3ʹ (reverse); U6 snRNA: 5′-CTCGCTTCGGCAGCACA-3′ (forward) and 5′-AACGCTTCACGAATTTGCGT-3 ʹ (reverse). The 2-ΔΔCt method was used to detect relative gene expression levels using U6 snRNA and GAPDH as internal controls.
Western Blot
Cells were lysed in RIPA buffer in supplement with protease inhibitor, and protein concentration quantification was processed using the BCA kit. About 40 μg protein samples were loaded onto 10% SDS-PAGE and transferred to a PVDF membrane. Non-fat milk was used to block the non-specific sites in the membrane, and then membranes were incubated with primary antibodies against NUP43 (ab235420) and GAPDH (ab181602, Abcam, Cambridge, MA, USA) at 4°C overnight, followed by 4 h of cultivation with the secondary antibody (ab6721, Abcam) at 25°C. Protein bands were visualized by BeyoECL Kit (Beyotime, Haimen, Jiangsu, China).
Bioinformatic Analysis
Expression levels of NCK1-AS1, miR-137, and NUP43 in GC tissues and normal tissues were analyzed with ENCORI. Moreover, the inter-connections of these three genes in GC tissues were also analyzed with ENCORI.
Cell Counting Kit-8 (CCK-8) Assay
Transfected cells were plated in a 96-well plate at the density of 2 × 103 cells/well. Ten μL CCK-8 reagent (Dojindo, Kumamoto, Japan) was added into each well and further incubated for 3 h. The absorbance of each well was measured at 450 nm with a microplate reader.
Colony Formation Assay
Six hundred cells were incubated in a 6-well plate in DMEM with 10% FBS for 14 days to allow colony formation. The medium was replaced every 3 days. Colonies were fixed with 4% paraformaldehyde and stained with 1% crystal violet. Images were captured and colony numbers were calculated.
Wound Healing Assay
Transfected cells were plated into a 6-well plate at the density of 1 × 106 cells/well and grown until about 90% confluence. A scratch was made on a cell monolayer with a pipette tip. Images were observed under an optical microscope and the migration distance was calculated after 48 h of incubation using Image J software.
Transwell Invasion Assay
A transwell chamber precoated with Matrigel (BD Biosciences, San Jose, CA, USA) was used to analyze cell invasion ability. Transfected cells (5 × 104) were harvested and, suspended in 200 μL serum-free medium, and plated to the upper chamber. DMEM containing 10% FBS was filled into the lower chamber. After incubation for 48 h in the described conditions, cells on the upper surface were wiped off by cotton swab. Meanwhile, cells that had invaded the lower chamber were fixed by polyformaldehyde for 10 min, stained with crystal violet for 15 min, and counted under an inverted microscope (IX83; Olympus, Tokyo, Japan).
Tumorigenesis Experiment
Animal experiments were conducted based on a protocol approved by the Animal Care and Use Committee of The Second Hospital of Shanxi Medical University and in accordance with the international standards-3R principle of animal welfare. For this, 3 × 106 cells with sh-NCK1-AS1 or sh-NC stable transfection were injected into the flank of nude mice (n = 5 per group). Width and length of the tumor were measured every 7 days, and volume was calculated using the formula length × (width)2/2. Tumor xenografts were grown for 28 days, and then sacrificed to excise the tumor tissues.
Immunohistochemistry (IHC) Assay
Excised tumors were fixed with 4% paraformaldehyde, and embedded in paraffin and sectioned at 4 μm thickness. Hydrogen peroxide at 3% was used to block the endogenous peroxidase activity for 20 min. Antigen was exposed in 10 mM citrate buffer (pH 6.0) by heat repair at 98°C for 2 min. Sections were incubated with primary anti-ki67 (ab92742, Abcam) overnight at 4°C and secondary antibody (ab6721, Abcam) for 20 min at room temperature. Immunoreactivity was visualized using 3,3′-diaminobenzidine (Sigma Chemical Co., St. Louis, MO, USA) and the sections were counterstained with hematoxylin.
Endogenous peroxidase activity was blocked by incubation in 3% hydrogen peroxide for 20 min. Antigen was exposed by heat repair in 10 mM citrate buffer (pH 6.0) at 98°C for 2 min. The sections were treated and incubated with primary anti-PRKCA (1:100 dilution; Abcam, Cambridge, UK) for 16 h at 4°C and horseradish peroxidase (HRP)-conjugated secondary antibody (Santa Cruz Biotechnology, CA, USA) for 20 min at room temperature. Immunoreactivity was visualized using 3,3ʹ-diaminobenzidine (Sigma Chemical Co., St. Louis, MO, USA) and sections were counterstained with hematoxylin and observed under a microscope.
RNA Immunoprecipitation (RIP) Assay
RIP assay was conducted with Millipore Magna RNA-binding Protein Immunoprecipitation Kit (Billerica, MA, USA) based on the user’s guide book to explore gene interactions. Cells were treated using RIP lysis buffer to collect cell lysates and then incubated with magnetic beads conjugated with anti-Ago2 or anti-IgG antibodies. Then, beads were collected and treated with Proteinase K and RNase-free DNase. TRIzol reagent was used to isolate RNA samples and subjected to RT-qPCR analysis to quantify NCK1-AS1, miR-137, and NUP43 expression levels.
Luciferase Reporter Assay
Interactions of miR-137 with NCK1-AS1 or NUP43 were analyzed with ENCORI. Wild-type (wt) or mutant (mt) sequences of NCK1-AS1 or NUP43 containing the miR-137 binding site were synthesized by GenePharma and inserted into the pmiR vector to generate wt/mt-NCK1-AS1/NUP43 luciferase constructs. Cells were co-transfected with luciferase vectors and synthetic miRNAs by Lipofectamine 2000. Dual-luciferase reporter system (Promega, Madison, WI, USA) was used to detect relative luciferase activity using Renilla luciferase activity as an internal control.
Statistical Analysis
Data from three independent experiments were analyzed with SPSS 19.0 and expressed as mean ± standard deviation. Statistical comparisons between two groups were analyzed with at-test, while those among three or more groups were analyzed with ANOVA and Tukey’s test. A P-value of less than 0.05 was considered as statistically significant.
Results
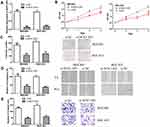
NCK1-AS1 Expression Level Was Upregulated in GC Tumor Tissues and Cells
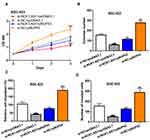
NCK1-AS1 expression level in GC was firstly analyzed at ENCORI and we found NCK1-AS1 expression was significantly elevated in GC tissues compared with normal tissues (Figure 1A). Moreover, we analyzed NCK1-AS1 expression in tissue samples we collected and we found that NCK1-AS1 expression was higher in GC tissues than in normal tissues (Figure 1B). In addition, we confirmed that NCK1-AS1 was more highly expressed in GC cells BGC-823 and MGC-803 in comparison with GES-1 cell (Figure 1C).
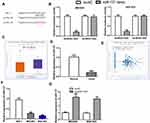
NCK1-AS1 Knockdown Inhibits GC Cell Proliferation, Colony Formation, Migration, and Invasion
Given the finding that NCK1-AS1 was highly expressed in GC, we knocked down NCK1-AS1 in GC cells using si-NCK1-AS1. An obviously downregulated expression level of NCK1-AS1 in GC cells transfected with si-NCK1-AS1 was observed (Figure 2A). Functionally, it was indicated that cell proliferation was efficiently suppressed by knockdown of NCK1-AS1 (Figure 2B), and that colony number declined in response to NCK1-AS1 downregulation, by comparing the colony numbers after growth for 2 weeks (Figure 2C). Meanwhile, the wound-healing assay showed the cell migration percentage of GC cells was significantly suppressed by knockdown of NCK1-AS1 by comparing the migration distance of GC cells at 0 h and 48 h after scratch creation (Figure 2D). Additionally, transwell invasion assay showed decreased NCK1-AS1 expression reduced the invasion ability of GC cells by comparing the invaded cell numbers in si-NCK1-AS1 and si-NC transfected groups (Figure 2E). These results indicated the depletion of NCK1-AS1 suppresses GC cell growth and motility in vitro.
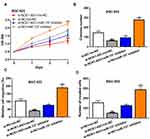
NCK1-AS1 Serves as a Sponge for miR-137
To explore the potential mechanisms of NCK1-AS1 in GC, miRNA targets were predicted by ENCORI and miR-137 was a putative target (Figure 3A and Supplementary Table 1). Dual-luciferase reporter assay showed miR-137 mimic transfection reduced luciferase activity of cells with wt-NCK1-AS1 but not mt-NCK1-AS1, which provides evidence for the interaction of NCK1-AS1 and miR-137 (Figure 3B). Subsequently, the expression level of miR-137 in GC was detected and the results indicated miR-137 level was lower in GC tissues than in normal tissues (Figure 3C and D). Meanwhile, a negative correlation of NCK1-AS1 and miR-137 was observed in GC tissues (Figure 3E). In addition, miR-137 was found to have decreased expression in GC cells compared with normal cells (Figure 3F). The silencing of NCK1-AS1 increased miR-137 expression level in GC cells (Figure 3G).
NCK1-AS1 Inhibits GC Cell Malignant Behaviors via Sponging miR-137
To explore whether miR-137 is involved in the NCK1-AS1 mediated stimulation effect on GC progression, GC cells were transfected with si-NCK1-AS1+miR-137 inhibitor, si-NCK1-AS1+mi-NC, si-NC+miR-137 inhibitor, or si-NC+mi-NC. Afterward, we found decreased miR-137 expression stimulated GC cell growth and motility (Figure 4A–D). In addition, the silence of miR-137 could partially reverse the inhibition effects of NCK1-AS1 on cell proliferation, colony formation, migration, and invasion (Figure 4A–D).
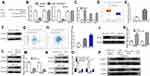
NCK1-AS1 Regulates NUP43 via miR-137
We found that NUP43 might be a putative miR-137 target through ENCORI analysis (Figure 5A and Supplementary Table 2). Dual-luciferase activity reporter revealed miR-137 mimic transfection reduced luciferase activity of wt-NUP43, while it did not influence mt-NUP43 in GC cells (Figure 5B). RIP assay indicated NCK1-AS1, miR-137, and NUP43 were co-enriched in anti-Ago2 pellets (Figure 5C). After that, we showed that NUP43 expression was elevated in GC tissues compared with in normal tissues (Figure 5D–F). Importantly, a negative correlation of miR-137 and NUP43 (Figure 5G), and a positive correlation of NCK1-AS1 and NUP43 (Figure 5H) was observed in GC tissues. Moreover, we found that NUP43 expression was increased in GC cells compared with normal cells, which is the same trend as displayed in GC tissues (Figure 5I and J). After that, expression analysis results showed NUP43 expression level can be increased by the miR-137 inhibitor (Figure 5K and L) and decreased by si-NCK1-AS1 (Figure 5M and N). Moreover, the stimulation effect of the miR-137 inhibitor on NUP43 expression can be restored by si-NCK1-AS1 (Figure 5O and P).
Silence of NCK1-AS1 Regulates GC Cell Behaviors via NUP43
We then explored whether NUP43 was a function target for NCK1-AS1 by transfecting si-NCK1-AS1+pNUP43, si-NC+pNUP43, si-NCK1-AS1+pcDNA3.1, and si-NC+pcDNA3.1 were transfected into GC cells. We found overexpression of NUP43 antagonized the inhibitory effects of NCK1-AS1 knockdown on GC cell proliferation, colony formation, migration, and invasion (Figure 6A–D).
Knockdown of NCK1-AS1 Inhibits GC Tumor Growth
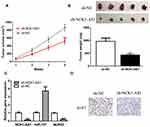
To explore the effects of NCK1-AS1 in vivo, cells with NCK1-AS1 knockdown were injected into mice and we showed both tumor volume and tumor weight were suppressed by NCK1-AS1 knockdown (Figure 7A and B). Furthermore, we showed NCK1-AS1 and NUP43 expression levels were decreased, whereas miR-137 expression level was increased in the sh-NCK1-AS1 group compared with the sh-NC group (Figure 7C). IHC assay showed ki-67 staining density was lower in the sh-NCK1-AS1 group than in the sh-NC group (Figure 7D).
Discussion
Emerging evidence indicated lncRNAs participate in various cellular processes to affect carcinogenesis.5,6 In GC, multiple lncRNAs were found to be abnormally expressed in GC through inhibiting or stimulating cellular malignant behaviors.14–16 Dai et al reported lncRNA MALAT1 contributes the response of GC cells to cisplatin via regulating PI3K/AKT pathway.14 Shi et al found that LINC00152 inhibits GC cell migration and invasion through alteration of the ERK/MAPK signaling pathway.15 Wei et al revealed that increased expression of lncRNA HOTAIR stimulates GC growth by upregulating COL5A1 via sponging miR-1277-5p.16 These studies uncovered the importance of lncRNAs in regulating GC progression.
NCK1-AS1 was found to serve as oncogenic lncRNA in several cancers.9,10,17 However, the roles and associated mechanisms of NCK1-AS1 in regulating GC remain unclear. In this study, we showed that NCK1-AS1 was upregulated expression in GC tissues and cells, indicating that NCK1-AS1 may play a crucial role in regulating GC progression. Subsequently, in vitro functional analyses indicated knockdown of NCK1-AS1 suppresses GC cell proliferation, colony formation, migration, and invasion. The conclusion of this work can be further strengthened by introducing si-NCK1-AS1 into a cell not expressing NCK1-AS1, which can provide evidence that the alternations of cell behaviors were due to the introduction of si-NCK1-AS1. After that, in vivo animal experiments showed that the silencing of NCK1-AS1 with sh-NCK1-AS1 could repress tumor growth compared with sh-NC.
As reported, lncRNA can serve as a sponge for miRNA to exert biological roles. To investigate miRNA molecules that may participate in the roles of NCK1-AS1, we predicted targets of NCK1-AS1 using the bioinformatic tool. miR-137 was selected for further analyses due to it ranking high among all predicted targets for NCK1-AS1, being negatively correlated with NCK1-AS1 expression, and previously being reported to serve a tumor suppressive role in several cancers.18–22 The interactions of NCK1-AS1 and miR-137 were validated by luciferase activity reporter assay and RIP assay. Thus, we further explored whether the impact of NCK1-AS1 in the development of GC was mediated by modulating miR-137. In this study, our results disclosed that NCK1-AS1 expedited proliferation, colony formation, migration, and invasion of GC cells, and the silence of miR-137 expression partially abolished the carcinogenic effect of NCK1-AS1. In other words, NCK1-AS1 contributes to GC tumor growth partially by targeting miR-361-3p.
miR-137 has been studied in cancers including triple-negative breast cancer, pituitary adenoma, and ovarian cancer to serve as a tumor suppressor.18–20 Consistent with these results, we also found that silencing of miR-137 can stimulate GC progression, suggesting a tumor suppressive role of miR-137. NUP43 was found highly expressed in luminal A and in HER2+ breast cancer, and could predict the poorer prognosis of cancer patients.23 Additionally, another work proved that NUP43 can be regulated by miR-409-5p to influence the response of chronic myeloid leukemia to imatinib.24 In this work, NUP43 was found to be negatively correlated with the expression of miR-137, and it had upregulated expression in GC tissues and cells. Rescue experiments indicated that NCK1-AS1 serves as ceRNA for miR-137 to upregulate NUP43 expression in GC cells to affect the aggressive behaviors of GC cells.
Conclusion
In conclusion, our results presented here showed, first, that NCK1-AS1 could promote GC cell proliferation, colony formation migration, and invasion in vitro. Also, it could regulate GC tumor growth in vivo. Second, NCK1-AS1 can regulate GC progression via the miR-137/NUP43 axis.
Author Contributions
All authors contributed to data analysis, drafting or revising the article, gave final approval of the version to be published, and agree to be accountable for all aspects of the work.
Disclosure
The authors report no conflicts of interest in this work.
References
1. Bray F, Ferlay J, Soerjomataram I, Siegel RL, Torre LA, Jemal A. Global cancer statistics 2018: GLOBOCAN estimates of incidence and mortality worldwide for 36 cancers in 185 countries. CA Cancer J Clin. 2018;68(6):394–424. doi:10.3322/caac.21492
2. Van Cutsem E, Sagaert X, Topal B, Haustermans K, Prenen H. Gastric cancer. Lancet. 2016;388(10060):2654–2664. doi:10.1016/S0140-6736(16)30354-3
3. ENCODE Project Consortium. An integrated encyclopedia of DNA elements in the human genome. Nature. 2012;489(7414):57–74. doi:10.1038/nature11247
4. Nagano T, Fraser P. No-nonsense functions for long noncoding RNAs. Cell. 2011;145(2):178–181. doi:10.1016/j.cell.2011.03.014
5. Batista PJ, Chang HY. Long noncoding RNAs: cellular address codes in development and disease. Cell. 2013;152(6):1298–1307. doi:10.1016/j.cell.2013.02.012
6. Wei L, Sun J, Zhang N, et al. Noncoding RNAs in gastric cancer: implications for drug resistance. Mol Cancer. 2020;19(1):62. doi:10.1186/s12943-020-01185-7
7. Yang J, Lian Y, Yang R, et al. Upregulation of lncRNA LINC00460 facilitates GC progression through epigenetically silencing CCNG2 by EZH2/LSD1 and indicates poor outcomes. Mol Ther Nucleic Acids. 2020;19:1164–1175. doi:10.1016/j.omtn.2019.12.041
8. Gong J, Wang Y, Shu C. LncRNA CHRF promotes cell invasion and migration via EMT in gastric cancer. Eur Rev Med Pharmacol Sci. 2020;24(3):1168–1176. doi:10.26355/eurrev_202002_20168
9. Chang H, Li B, Zhang X, Meng X. NCK1-AS1 promotes NCK1 expression to facilitate tumorigenesis and chemo-resistance in ovarian cancer. Biochem Biophys Res Commun. 2020;522(2):292–299. doi:10.1016/j.bbrc.2019.11.014
10. Qiao Z, Dai H, Zhang Y, Li Q, Zhao M, Yue T. LncRNA NCK1-AS1 promotes cancer cell proliferation and increase cell stemness in urinary bladder cancer patients by downregulating miR-143. Cancer Manag Res. 2020;(12):1661–1668.
11. Kloosterman WP, Plasterk RH. The diverse functions of microRNAs in animal development and disease. Dev Cell. 2006;11(4):441–450. doi:10.1016/j.devcel.2006.09.009
12. Shin VY, Chu KM. MiRNA as potential biomarkers and therapeutic targets for gastric cancer. World J Gastroenterol. 2014;20(30):10432–10439. doi:10.3748/wjg.v20.i30.10432
13. Salmena L, Poliseno L, Tay Y, Kats L, Pandolfi PP. A ceRNA hypothesis: the Rosetta Stone of a hidden RNA language? Cell. 2011;146(3):353–358. doi:10.1016/j.cell.2011.07.014
14. Dai Q, Zhang T, Li C. LncRNA MALAT1 regulates the cell proliferation and cisplatin resistance in gastric cancer via PI3K/AKT pathway. Cancer Manag Res. 2020;(12):1929–1939.
15. Shi Y, Sun H. Down-regulation of lncRNA LINC00152 suppresses gastric cancer cell migration and invasion through inhibition of the ERK/MAPK signaling pathway. Onco Targets Ther. 2020;13:2115–2124. doi:10.2147/OTT.S217452
16. Wei Z, Chen L, Meng L, Han W, Huang L, Xu A. LncRNA HOTAIR promotes the growth and metastasis of gastric cancer by sponging miR-1277-5p and upregulating COL5A1. Gastric Cancer. 2020. doi:10.1007/s10120-020-01091-3
17. Huang L, Li X, Ye H, et al. Long non-coding RNA NCK1-AS1 promotes the tumorigenesis of glioma through sponging microRNA-138-2-3p and activating the TRIM24/Wnt/β-catenin axis. J Exp Clin Cancer Res. 2020;39(1):63. doi:10.1186/s13046-020-01567-1
18. Lee SJ, Jeong JH, Kang SH, et al. MicroRNA-137 inhibits cancer progression by targeting Del-1 in triple-negative breast cancer cells. Int J Mol Sci. 2019;20(24):E6162. doi:10.3390/ijms20246162
19. Duan J, Lu G, Li Y, Zhou S, Zhou D, Tao H. miR-137 functions as a tumor suppressor gene in pituitary adenoma by targeting AKT2. Int J Clin Exp Pathol. 2019;12(5):1557–1564.
20. Song R, Liu Z, Lu L, Liu F, Zhang B. Long noncoding RNA SCAMP1 targets miR-137/CXCL12 axis to boost cell invasion and angiogenesis in ovarian cancer. DNA Cell Biol. 2020;39(6):1041–1050. doi:10.1089/dna.2019.5312
21. Chen W, Du J, Li X, Zhi Z, Jiang S. microRNA-137 downregulates MCL1 in ovarian cancer cells and mediates cisplatin-induced apoptosis. Pharmacogenomics. 2020;21(3):195–207. doi:10.2217/pgs-2019-0122
22. Hao Y, Li X, Chen H, Huo H, Liu Z, Chai E. Over-expression of long noncoding RNA HOTAIRM1 promotes cell proliferation and invasion in human glioblastoma by up-regulating SP1 via sponging miR-137. Neuroreport. 2020;31(2):109–117. doi:10.1097/WNR.0000000000001380
23. Tian C, Zhou S, Yi C. High NUP43 expression might independently predict poor overall survival in luminal A and in HER2+ breast cancer. Future Oncol. 2018;14(15):1431–1442. doi:10.2217/fon-2017-0690
24. Liu YY, Jiao WY, Li T, Bao YY. MiRNA-409-5p dysregulation promotes imatinib resistance and disease progression in children with chronic myeloid leukemia. Eur Rev Med Pharmacol Sci. 2019;23(19):8468–8475. doi:10.26355/eurrev_201910_19159
 © 2020 The Author(s). This work is published and licensed by Dove Medical Press Limited. The full terms of this license are available at https://www.dovepress.com/terms.php and incorporate the Creative Commons Attribution - Non Commercial (unported, v3.0) License.
By accessing the work you hereby accept the Terms. Non-commercial uses of the work are permitted without any further permission from Dove Medical Press Limited, provided the work is properly attributed. For permission for commercial use of this work, please see paragraphs 4.2 and 5 of our Terms.
© 2020 The Author(s). This work is published and licensed by Dove Medical Press Limited. The full terms of this license are available at https://www.dovepress.com/terms.php and incorporate the Creative Commons Attribution - Non Commercial (unported, v3.0) License.
By accessing the work you hereby accept the Terms. Non-commercial uses of the work are permitted without any further permission from Dove Medical Press Limited, provided the work is properly attributed. For permission for commercial use of this work, please see paragraphs 4.2 and 5 of our Terms.